Data Model
Introduction
HAWK’s data model is designed accommodate wildlife health data from various sources, including local to national wildlife health surveillance systems, research initiatives, and citizen-science based projects.
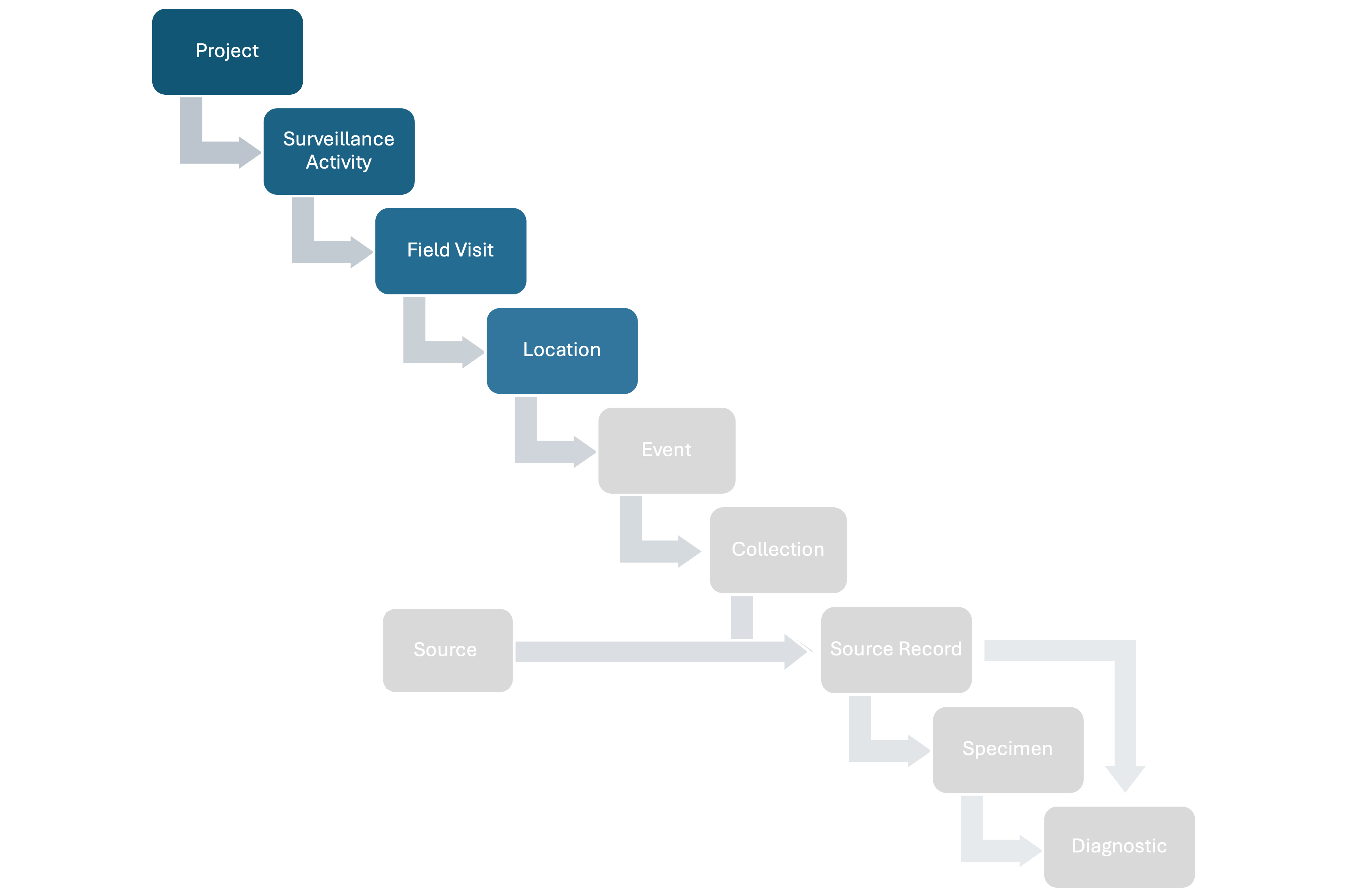
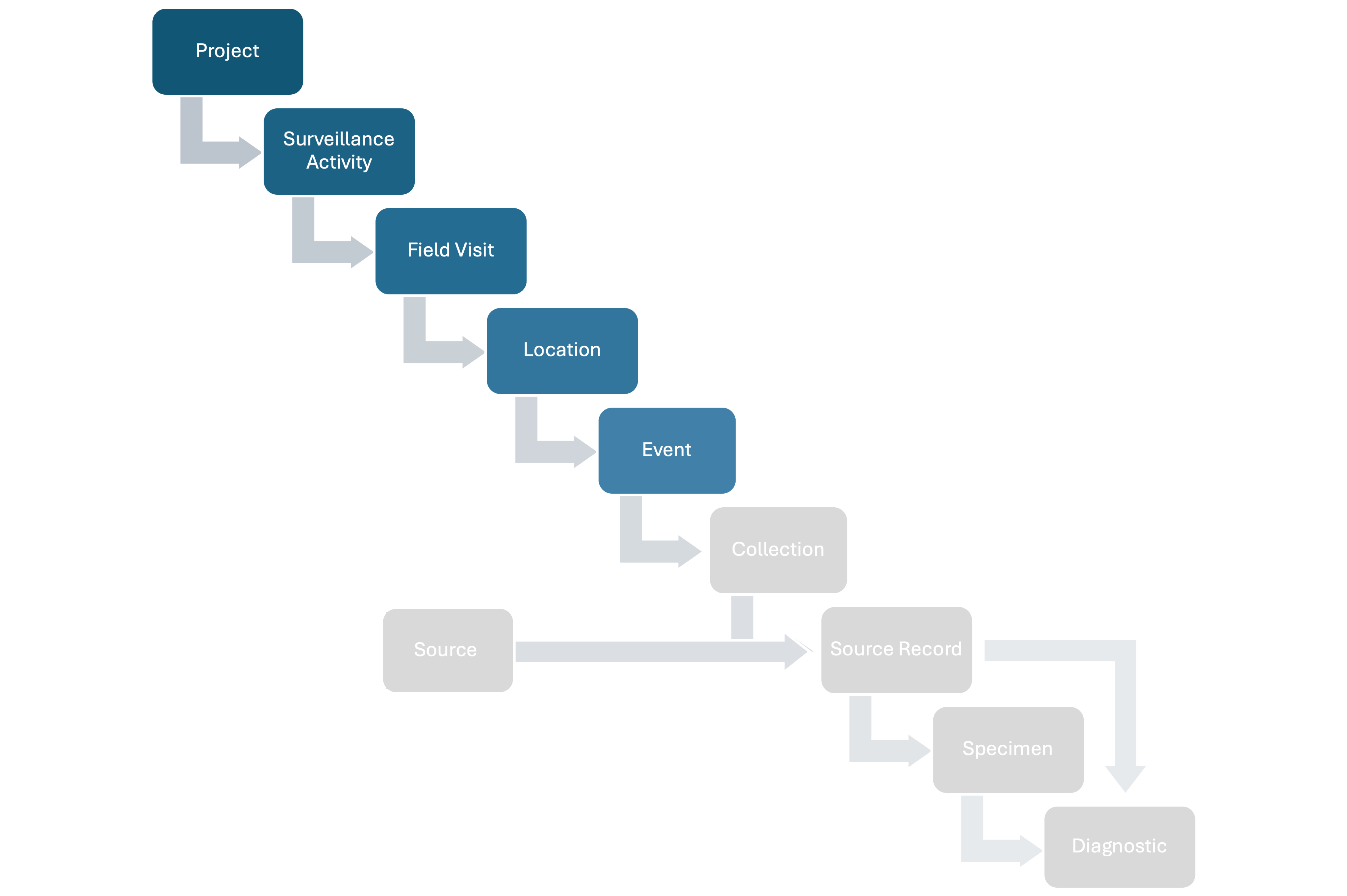
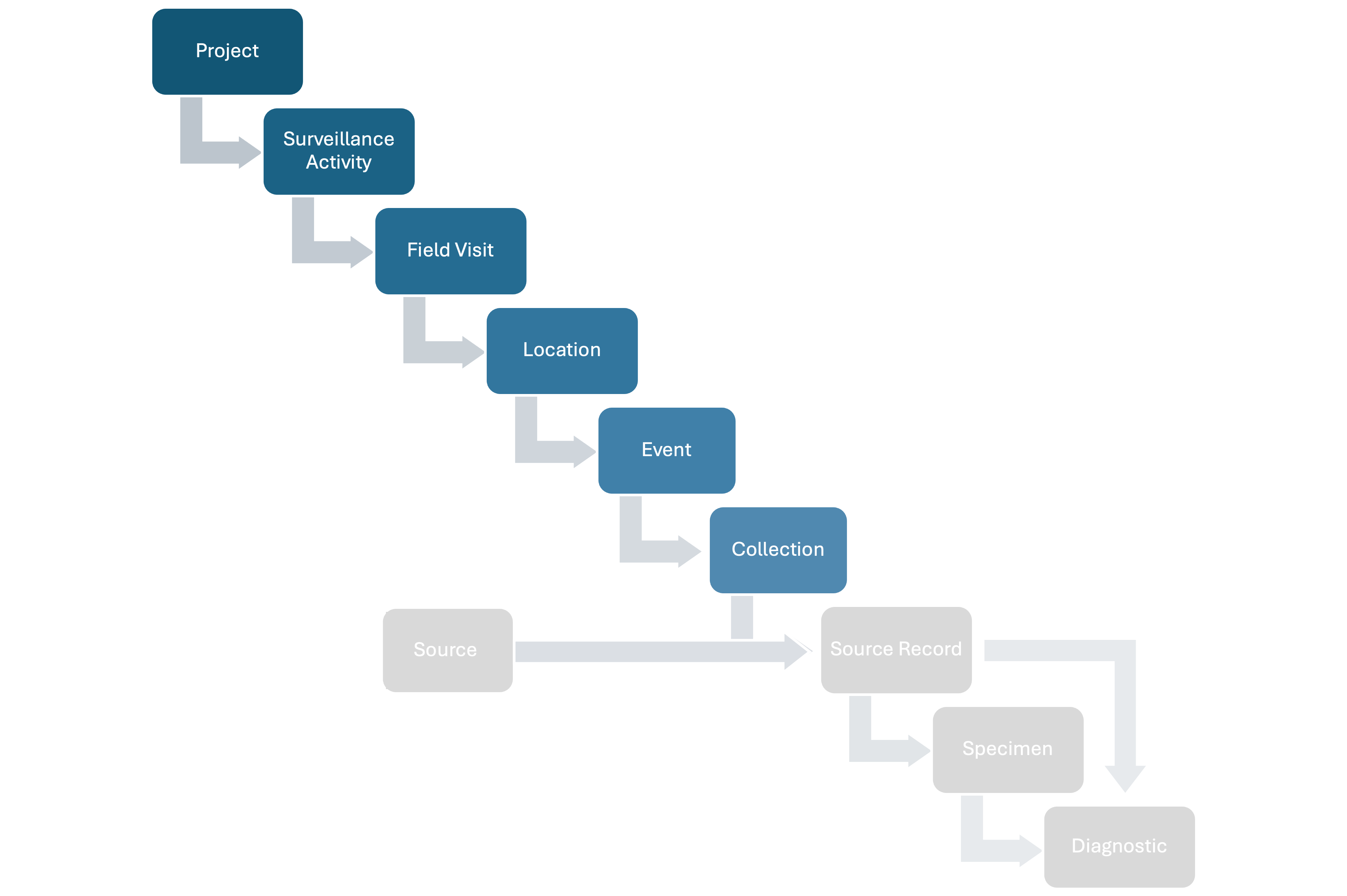
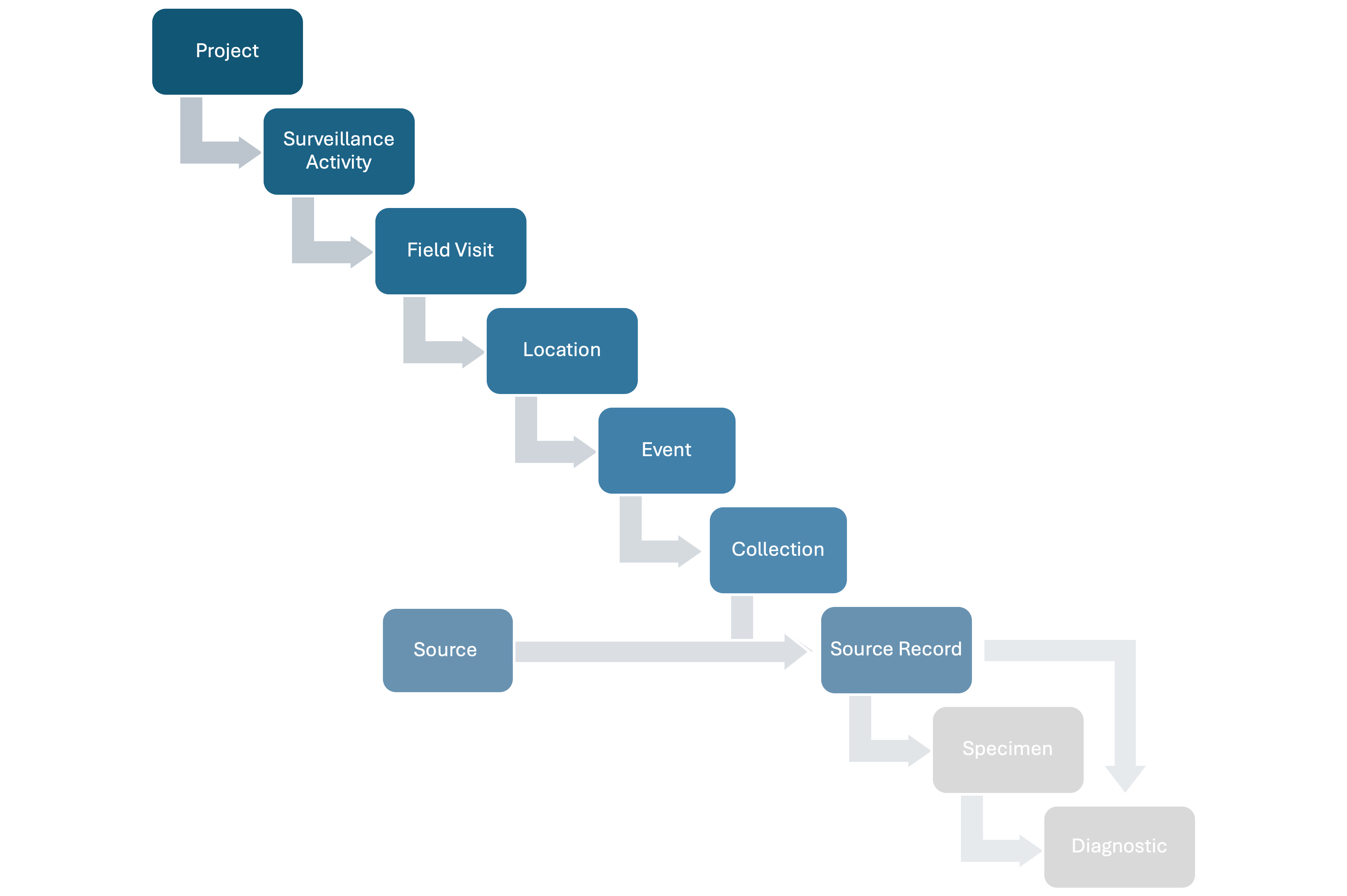
HAWK structures wildlife health surveillance data following a hierarchy, capturing relevant epidemiological information at each step. The full hierarchy includes:
- Projects: A set of Surveillance Activities with a common leader and organizer (e.g., a national wildlife health surveillance system).
- Surveillance Activities: Specific sets of surveillance goals and methodology (who, where, when, what, why, how) documented following a standard metadata format.
- Field Visits: Time period during which activities are conducted in the field.
- Locations: Specific areas where activities are conducted.
- Collection Areas and Collection Points: Effort units associated with the generation of data in a explored area (e.g., sector within protected area) and in a linear feature (e.g., transect or ranger patrol track), respectively.
- Events: Point epidemiological units with a spatiotemporal coordinate where wildlife health surveillance data is collected.
- Point Collection: Effort units associated with the generation of data in an Event (e.g., a set of mist nests placed during a specific period of time).
- Sources: Units that can provide Specimens for hazard detection or whose health is assessed (observed-only) at a specific time. There are four Source types:
- Group: groups of animals of the same species.
- Animal: individual animals
- Environmental: environmental sampling sites (e.g., ponds or feces in the field).
- Arthropod: arthropod capture sites.
- Source Records: Records of the Sources at a specific time t.
- Specimens: Tissues taken from Source Records.
- Diagnostics: Tests conducted on Sources or collected Specimens to assess health hazards.
- Results: The result(s) of a Diagnostic for the targeted hazard(s) and the corresponding interpretation for each hazard.
- Interpretation: The final status assigned to a Specimen or Source Record following documented case definitions.
The basic relationships among the basic units of the data model are shown in Figure 1 below:
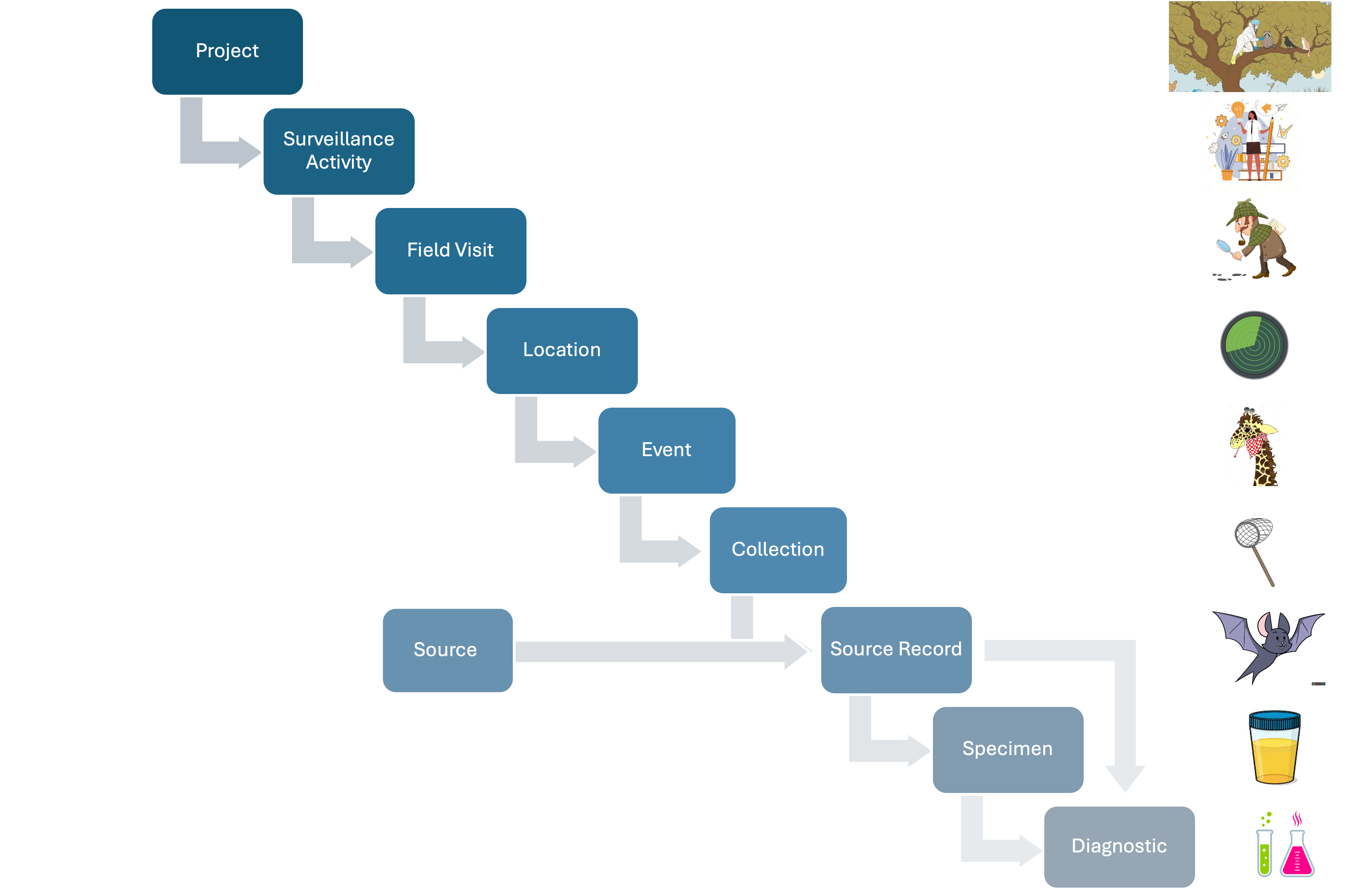
Figure 1: Basic relationships among the basic units of the data model.
HAWK employs a modular approach to the data model, enabling components to be added based on the specific WHS needs. When data comes from wildlife mortality reported by a community member through a mobile application:
- The Surveillance Activity contains a description of the methodology.
- The Field Visit could be defined annually to categorize reports by year.
- The Location could be a county or zip code.
- An Event is the epidemiological unit with latitude, longitude, and time representing the position where dead animals found.
- The Animal Source Records describe the dead animals.
- No Specimens, Diagnostics, or Laboratory data are generated.
- No Collection effort is linked to the Events and the animals are found opportunistically.
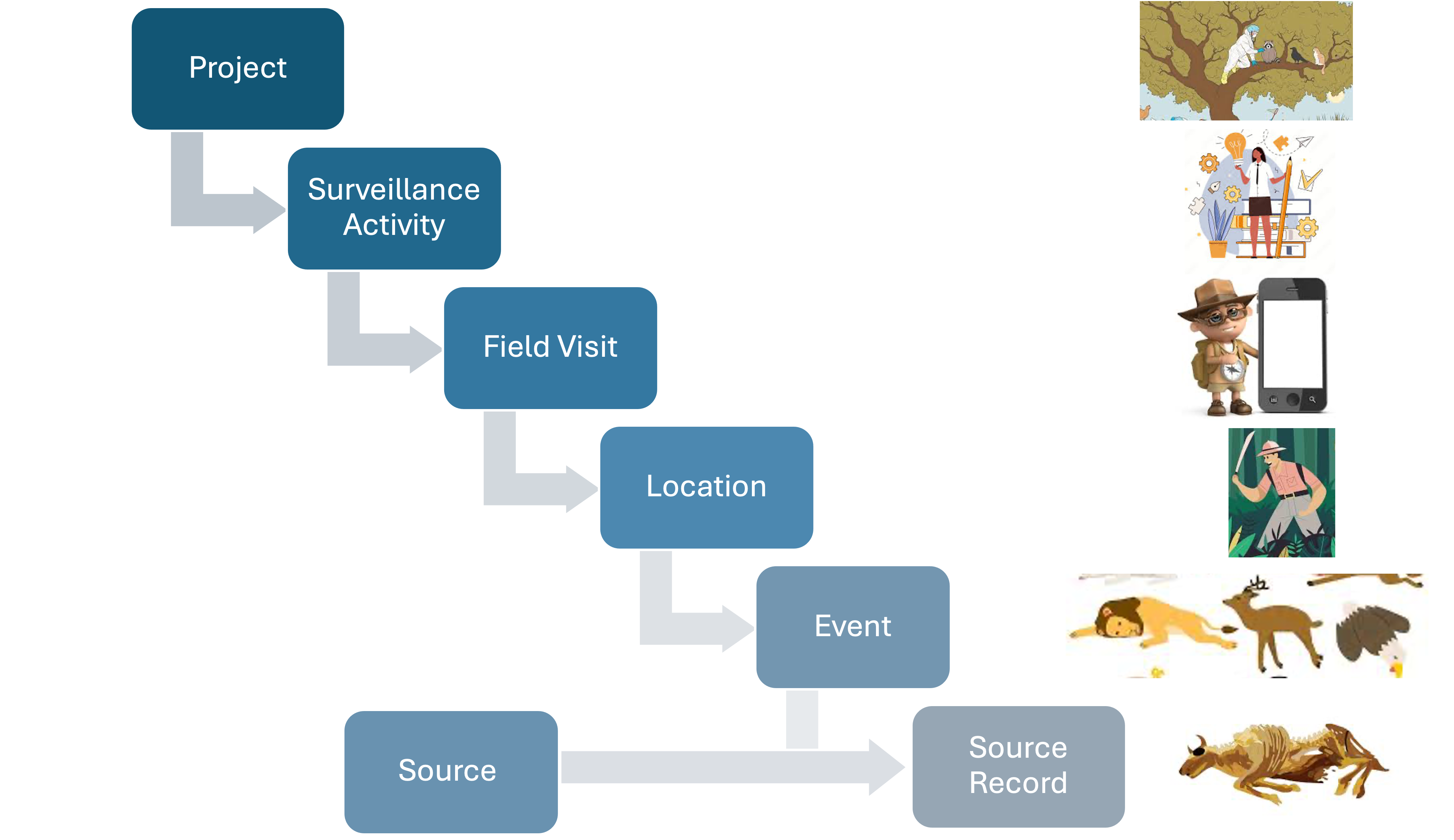
Figure 2: Components of the data model to contain data from example 1.
This flexible structure ensures that the model remains efficient and scalable, adapting to different surveillance needs.
The data model is not designed for wildlife population monitoring. However, it includes key identifiers that enable seamless integration between wildlife health and population data.
More complex relationships between the fundamental units of the data model, as well as additional details, are covered in the Complexities section. Before exploring these advanced topics, it is recommended to first understand the core components of the data model, from Project to Diagnosis, Results, and Interpretation, by continuing with this documentation. Use the menu in the top-right corner of this website to navigate to the relevant sections.
Main Units of the Data Model
Project
A Project in the data model represents a surveillance initiative supported by specific entities and lead by specific individuals. Examples include the PREDICT Project funded by USAID, a single cross-sectional study with one field visit to a specific location (e.g., sample collection in a market), or a national or local wildlife health surveillance network managed by a government agency.
Projects serve as the highest hierarchical unit in the database and must contain at least one Surveillance Activity. They can be time-limited, spanning a single date, or ongoing for an extended period as needed.
Key properties of a Project include:
- Project ID
- Project Code
- Project Cross-Reference ID
- Project Cross-Reference Origin
- Project Leader
- Project Funding Source
Other attributes are provided in the Data Dictionary
Surveillance Activity
A Surveillance Activity in the data model refers to a coordinated set of activities aimed at detect diseases, demonstrate disease freedom, measure incidence or prevalence, assess trends in specific health and health hazards within defined populations. In general, a specific set of methods, strategies, and objectives should equal a specific Surveillance Activity.
For example, the longitudinal assessment of coronavirus shedding in two Eidolon helvum bat roosts in Africa involved collecting a fixed number of fecal samples from two bat roosts of the same species every month for 12 months, with subsequent testing for Coronaviridae sp.
Each Surveillance Activity includes detailed metadata describing its objectives and methods, such as:
- Targeted species, populations, and hazards
- Samples and collection methods
- Diagnostics conducted and case definitions
A Surveillance Activity can span multiple Field Visits, Locations, and Source types (e.g., Groups, Animals, Environmental and Arthropod Sources).
Generally, a Surveillance Activity usually includes Field Visits, Locations, Events, Sources, Source Records, and Diagnostics. However, exceptions exist (see Complexities section).
Most Surveillance Activity properties align with standard documentation of surveillance methods, including:
- Start and End Dates
- Targeted Hazards and Taxa
- Involved partners
- Definitions of Locations, Events, and Sources
- Source identification, Specimens, and Collection methods
- Diagnostic techniques and case definitions
Outbreak Investigation
Each outbreak investigation is considered a Surveillance Activity. Outbreak Surveillance Activities can encompass the initial Field Visit that led to the outbreak detection and subsequent Field Visits associated with the investigation and control. For instance, an outbreak may first be detected during routine patrols by rangers. In such cases, the initial Field Visit and its components should be belong to both the ranger patrol Surveillance Activity and also the Outbreak Surveillance Activity. Follow-up Field Visits (e.g., by veterinarians or public health officers investigating the outbreak) should be assigned exclusively to the Outbreak Surveillance Activity (see Complexities section).
Field Visit
A Field Visit in the data model represents a defined time period—including a start and end date—during which Locations are visited, Events and Sources are identified and observed, and Specimens might be collected and documented.
The length of a Field Visit is flexible. Key properties of a Field Visit include:
- Start and End Date
- Field Visit ID and Code
- Field Visit Cross-Reference ID and Origin
- Field Visit Leader
If there are a Field Visit attributes that are not part of the current data model but are of interest to track, they must be reported in the Surveillance Activity metadata. It is recommended to maintain such additional variables (e.g., personnel during the Field Visit) in a separate system, such as another database or an Excel sheet, for reference. Common extra Field Visit attributes can be added to the data model in the future. Missing options for single- and multi-selection attributes of Field Visits can be added as long as they maintain a controlled vocabulary to ensure consistency and data integrity.
Location
In the data model, a Location represents a general area where Events can be identified. Unlike Events, which are defined by exact latitude and longitude coordinates, Locations serve as a broader spatial grouping for organizing data. A single Location can contain zero Events (e.g., when an area is surveyed but no Event is recorded) or multiple Events (e.g., multiple observations or findings within the same area).
The meaning of a Location depends on the methodology of the Surveillance Activity. For Arthropod collection, a Location may represent a parcel where traps are set. In a structured, hierarchical study, a Location could correspond to a grid cell. During ranger patrols, a Location might be defined as the entire protected area or a specific zone within it. Users must define the specific unit a Location represents and document it in the Surveillance Activity metadata. We strongly suggest to define a Location as the area unit of interest that is explored to find Events (e.g., area covered in a beach on the search of dead birds) or the smallest area unit containing Events (e.g., a grid cell where traps are placed).
Key attributes of a Location include:
- Location ID and Code
- Location Cross-Reference ID and Origin
- Location Type
If there are a Location attributes that are not part of the current data model but are of interest to track, for example, specific environmental features, they must be reported in the Surveillance Activity metadata. It is recommended to maintain such additional attributes in a separate system, such as another database or an Excel sheet, for reference. Common extra Location attributes can be added to the data model in the future. Missing options for single- and multi-selection attributes of Location can be added as long as they maintain a controlled vocabulary to ensure consistency and data integrity.
Collection Area
In the data model, Collection contains the information associated with the effort to obtain Sources in the field actively. In this case, Collection Area refers to the effort to find or document Events in an area during a specific period of time. For example, the surface covered in the search of dead birds, and the amount of time spent covering that area. The Collection area can be equivalent to the area of the Location (one Collection Area in the Location) or they can be smaller units within the Location (e.g., the Location is a full grid cell and each grid cell is explored with a different effort). Sources observed or sampled opportunistically are not tied to an effort, therefore, the Collection Area units do not apply.
The meaning of a Collection Area depends on the methodology of the Surveillance Activity. For an outbreak, dead animals are actively searched for in a beach. The full beach can represent a single Collection Area within the beach (the Location) or multiple areas within the beach (the Location) can each represent a Collection Area.
Key attributes of a Collection Area include:
- The spatial effort unit
- The spatial effort (area covered)
- The temporal effort unit
- The temporal effort (period of time to cover the area)
- The spatial coordinates of the area
If there are a Collection Area attributes that are not part of the current data model but are of interest to track, for example, specific environmental features, they must be reported in the Surveillance Activity metadata. It is recommended to maintain such additional attributes in a separate system, such as another database or an Excel sheet, for reference. Common extra Collection Area attributes can be added to the data model in the future. Missing options for single- and multi-selection attributes of Collection Area can be added as long as they maintain a controlled vocabulary to ensure consistency and data integrity.
Collection Track
A Collection Track refers to the effort to find or document Events in linear features within a Location during a specific period of time. For example, the distance travelled by rangers during a conducted in a protected area where they find dead animals or a population monitoring transect. The effort also includes and the amount of time to cover the track. A Location can contain one or multiple Collection Tracks. Sources observed or sampled opportunistically are not tied to an effort, therefore, the Collection Track units do not apply.
Collection Tracks are not nested in Collection Areas but both are nested under Locations. If a methodology involves Events nested in Collection Tracks and these tracks nested in Collection Areas then, the link between both types of Collection can be made through the spatial data and through Tags (see Complexities).
Key attributes of a Collection Track include:
- The spatial effort unit
- The spatial effort (distance travelled)
- The temporal effort unit
- The temporal effort (period of time to travel the reported distance)
- The spatial coordinates of the track
If there are a Collection Track attributes that are not part of the current data model but are of interest to track, for example, specific environmental features, they must be reported in the Surveillance Activity metadata. It is recommended to maintain such additional attributes in a separate system, such as another database or an Excel sheet, for reference. Common extra Collection Tracks attributes can be added to the data model in the future. Missing options for single- and multi-selection attributes of Collection Tracks can be added as long as they maintain a controlled vocabulary to ensure consistency and data integrity.
Event
In the data model, an Event represents a distinct wildlife health event recorded at a specific longitude, latitude, and timestamp. Each Event is a point that can be associated with no active effort to collect data from the field up to an unlimited number of efforts (Collection Points).
The definition of a “wildlife health event”, and therefore, what an Event represents in a Surveillance Activity will inevitably vary depending on the main epidemiological unit of interest receiving the longitude, latitude, and temporal stamp. Examples include:
- A site where Animals are captured at time t to obtain Specimens
- The position and time a dead animal is found during a ranger patrol
- A position and time where water from a pond is sampled for analysis
- A site where one or multiple traps for mosquitoes are deployed
Depending on the methodology employed, an Event can contain a single trap and, therefore, a single Collection Point, or it can contain multiple traps. In this case, the design implies assigning the same longitude and latitude to a set of traps and then each one of them could be recorded as individual Collection Points.
In the case of beached fish, an Event can represent the position and time of recording of each individual dead fish in one extreme, or the total count of dead fish across the beach reported as a single point in the other extreme. In the context of a wet market, the definition of Event can be applied to the market, to vendors, to the stalls of vendors, or to the cages in the stalls at time t. For study A, the Event could be the grid cell where traps are deployed, whilst for study B it could represent each trap within a grid cell.
When the collection of data from animals or sampling sites is opportunistic (no effort involved), for example, a dead animal found during a ranger patrol, then the position of that animal represents the Event.
In active surveillance activities, an Event might not have any data besides the effort, for example, when no animals are captured at time t.
An Event can include or not healthy animals. In the example of the dead animal found by the ranger at time t, it could be possible that the Event definition includes the documentation of healthy animals at the same position.
An Event can include more than one Source type. For example, mosquito traps (Arthropods) and Environmental sampling under the same Event.
It is possible that specific latitude and longitude coordinates are not of interest in a Surveillance Activity. For example, a set of mosquito tramps deployed in parcels whose specific spatial position within each parcel is not relevant, and the parcels are visited multiple times. In this case, the suggestion is to consider each parcel as a Location, the Event is a spatiotemporal point within the parcel area, and the traps are documented as specific Collection Points or as a single Collection Point depending on the granularity desired.
This flexibility allows HAWK to accommodate diverse surveillance strategies, ensuring meaningful and context-appropriate data representation.
It is up to the user to define if the Event is a unit of interest and what it represents, and to document this definition in the Surveillance Activity metadata. To ensure consistency, each Surveillance Activity must maintain a well-establish Event definition. We strongly suggest that an Event represents the minimal unit where Sources Records are contained in, for example, capturing, containing, or sampling points (mist-nets, cages, traps, position where water is obtained) or the position of dead animals.
Key attributes of a Event include:
- Start Date
- Event ID
- Event Code
- Event Cross-Reference ID
- Event Cross-Reference Origin
- Longitude & Latitude
- Coordinate Reference System (CRS) used
If there are Event attributes that are not part of the current data model but are of interest to track, they must be reported in the Surveillance Activity metadata. It is recommended to maintain such additional attributes in a separate system, such as another database or an Excel sheet, for reference. Common extra Event attributes can be added to the data model in the future. Missing options for single- and multi-selection attributes of Event can be added as long as they maintain a controlled vocabulary to ensure consistency and data integrity.
When an Event encompasses Source Records that are recorded under the same point but they were actually distributed across an area, it is possible to add the area around the Event that encompassed these findings, for example, dead birds in a radius of 20 meters recorded as a single Event.
Collection Point
In the data model, a Collection Point represents the effort involved in observing, detecting, capturing, or otherwise identifying Sources at an Event.
Examples of Collection Point include:
- The time spent by an observer at a fixed position to identify dead birds in a wetland.
- The camera traps deployed at a specific site to photograph animals and the hours they were active.
- The traps deployed during a period of time to obtain mosquitoes.
Collection Points also allow to characterize problems during the search for Sources, such as camera traps running out of battery or stolen, torn mist nets, etc.
A Collection Point can contain between zero and an unlimited number of Source Records. For example, a capture effort that results in no animals caught.
An Event might not include any Collection Point. Examples include Sources found opportunistically (e.g., dead birds accidentally found in a beach) or sampling of the animals in a wet market.
A Collection Point typically contains a specific type of Source Record. This is because observations, capturing, and sampling efforts generally focus on a specific type of target, whether Group, Animal, Environmental, or Arthropods. However, a single Event can include multiple Collection Points targeting different Specimen Sources. For example, mosquito traps and an air filtration device can be linked to the same Event.
Collections are defined by:
- The spatial effort (e.g., “number of mist nets”, “number of CO₂ traps”, “number of Camera traps”, “distance walked”; and “area scanned”).
- The temporal effort (e.g., “hour deployed”).
When Source Records are not linked to a Collection Point (e.g., opportunist finding of dead animals), then the *Collection Point** is “NA”.
If there are Collection Point attributes that are not part of the current data model but are of interest to track, they must be reported in the Surveillance Activity metadata. It is recommended to maintain such additional attributes in a separate system, such as another database or an Excel sheet, for reference. Common extra Collection Points attributes can be added to the data model in the future. Missing options for single- and multi-selection attributes of Collection Points can be added as long as they maintain a controlled vocabulary to ensure consistency and data integrity.
Source and Source Records
General Overview
In the data model, a Source is a unit of interest that can be observed, provide Specimens for analysis or that can be observed-only. The data model manages four types of Sources: i) a co-specific group of animals (Group), ii) individual animals (Animal), iii) sites where arthropods can be obtained from (Arthropod), and iv) sites where biotic and abiotic tissue not linked to any other type of Source can be obtained from (Environmental). A Group Source could correspond to bats roosting at cave X sourcing guano, an Animal Source could be a collared animal, an Arthropod Source could be a site where mosquito larvae are collected from, and an Environmental Source could be a pond where water, air, and sediment are collected from or feces found in the field whose animal of origin is unknown. Sources contain time-independent data only, such as the species of an animal in the case of a Group or Animal Sources (more below).
All Sources can potentially be identified and tracked over time if needed. However, there are limitations, Environmental and Arthropod Sources are site-specific, making them easier to track across different time points. Animal and Group Sources may not always be individually identifiable, which can prevent tracking them over time.
**A Source collected, or captured at time t can be linked to Events through a Point Collection or not (see previous section) via a Source Record, which represents the Source sampled or observed at a specific time t. Source Records contain only time-dependent data, such as health status at time t for Group and Animal Sources or the number of arthropods trapped at time t* for Arthropod Sources.
Individually identified Sources can tracked across multiple Events, even across different Surveillance Activities. For example, a bat (Source) captured in a mist net (Collection) deployed at a roost at time t (Event), individually identified, and later recaptured in another Event t′. This bat is linked to both the original capture Event and the recapture Event, with a unique Source Record for each occurrence.
Sources that are not identified will be linked as a Source Record to the Event where they were observed or captured only. For example, a bat (Source) that is captured (Collection Point) and sampled but not marked or a dead animal not identified found by a ranger during a patrol. The bat and the animal found are linked to a single Event, the one they were capture or observed.
Sources found opportunistically are still linked to an Event, even if no effort is involved. For example, a member of the public reports a dead animal (Source Record) found at time t (Source Record) at a beach (Event).
Sources Records can provide between zero (e.g., observation only) up to indefinite number of Specimens to conduct Diagnostic tests.
Group Source
A Group Source is a unit of co-specific individuals (animals of the same species) that belong to a common entity, such as a herd, roost, area, site, farm, cage, stall, or enclosure, forming a single epidemiological unit. Group Sources can be observed, captured, diagnosed as a full unit, and provide Specimens collectively at a given time t when the Event occurs.
Key attributes of a Group Source include:
- Species
- Group Source ID
- Group Source Cross Reference ID
- Collection Cross Reference Origin
The primary purpose of a Group Source is to record individuals at the species level rather than tracking each animal individually. This approach is useful when herds are the unit of interest, when protected area rangers document multiple animals of the same species during a health event, or when animals in a market are grouped in cages or stalls without individual identification. For example, if a single cage contains animals from two different species and they are not tracked individually, they can be recorded as two separate Group Sources—one per species.
Group Source Record
A Group Source Record documents the animals within a Group Source at time t, categorized by sex, age, and health status (e.g., healthy, injured, sick, or dead). Additional attributes include:
- Observed anomalies
- Potential cause of disease
- Potential cause of death
Since properties of a Group Source Record are recorded at the group level, multiple options can be reported for these attributes. For example, if a Group Source Record includes three dead animals, several potential causes of death may be listed. However, it is not possible to determine how these potential causes or other properties are distributed across individuals within the Group Source Record—only that they were present in at least one individual.
A Group Source Record can include a mix of dead, diseased, injured, and healthy individuals of the same species, or it may consist solely of one category if all individuals share the same status (e.g., all are dead). Additionally, even a Group Source Record consisting only of healthy animals - only can be part of a Health Event, depending on the Event’s definition.
A Group Source Record may contain a single individual. For example, rangers patrolling a protected area might find one dead animal of species X and two dead animals of species Y at the same site (Event). The animals can be documented as distinct Group Source Records (one group per species).
An Event can include multiple *Group Source Records of the same species. For example, if a vendor at a market keeps animals of the same species in two separate cages, and the Event is a vendor, then each cage could be considered a distinct unit. Consequently, there would be two Group Source Records for the same species under the same vendor (i.e., within the same Event).
A Group Source can be directly used for a Diagnostic (e.g., assessing the body condition of a specific herd using a standardized metric). The data model supports Diagnostics applied to an entire group rather than just a Specimen taken from that Group Source. Specimens from Group Sources can be stored, exported, and used for Diagnostics. On the contrary,Group Source cannot have a collective necropsy. This is because their collection and handling present an opportunity to gather data at the individual level and recorded as an Animal Source. Carcasses of animals of a Group Source collected or taken for Necropsy must be converted into Animal Sources. These Animal Sources are recorded as former members of the Group Source (see the Complexities section for further details).
For similar reasons, Group Sources cannot be removed from the field. If dead animals from a Group Source are collected, each must be documented as an Animal Sources originating from the corresponding Group Source whose Carcass is taken (see Animal Sources and Carcass below).
The database does not accept live animals from Group Sources be taken ex-situ.
An animal of species X recorded in a Group Source Record must not be duplicated as an Animal Source Record, and vice versa. If an Event contains both a Group Source Record and an Animal Source Record of species X, the total number of animals of that species in the Event is the sum of those in the Group Source Record and the individual(s) recorded separately as Animal Source Record (e.g., the individuals in a group of animals of species X plus a carcass collected of the same species that would have belonged to the count of dead animals of the Group Source Record if not collected).
For example, consider a herd of 20 cows illegally raised in a protected area whose health is assessed at time t. If two cows are sampled, the two sampled cows should be documented as two separate Animal Source Records (one per cow), while the remaining 18 cows are recorded as a single Group Source Record, categorized by sex, age, and health status. In this scenario, the total number of cows is 20—the 18 recorded in the Group Source Record plus the two documented as Animal Source Records. The “herd” identity of these 20 cows split in three Source Records can be maintained using a cluster (see Complexities). If, however, the Group Source Record incorrectly includes all 20 cows while two of them are also recorded as Animal Source Record, the total count would incorrectly sum to 22.
Animal Source
An Animal Source represents a single individual. Animal Sources can be observed, captured, tested, necropsied, and provide Specimens at time t, directly or during a Necropsy.
Key attributes of a Animal Source include:
- Species
- Animal Source ID
- Animal Source Cross Reference ID
- Sex
For example, in a live animal market, if data is collected at the individual level, each animal—whether of the same or different species—must be recorded as a separate Animal Source. In contrast, if animals are documented collectively (e.g., by species in a shared cage), they would be classified under a Group Source rather than as individual Animal Sources.
Any past marking codes assigned of the animal are considered immutable and are recorded as part of the Animal Source data.
Animal Source Record
An Animal Source Record represents an individual Animal Source at time t.
Key attributes of a Animal Source Record include:
- Age
- Observed Health Status
- Observed Anomalies
- Potential Causes of Disease or Death
Some properties of an Animal Source Record allow single or multiple selections. For instance, an animal’s health status is recorded as one category (e.g., “live healthy” or “live sick”), whereas multiple anomalies may be reported for the same individual at time t (e.g., wounds, hair loss, diarrhea).
An Animal Source Record can be categorized as dead, diseased, injured, or healthy at time t. A live, healthy Animal Source Record can still be part of an Event, depending on the Event’s definition (e.g., an apparent healthy animal of species X observed near dead animals of species X, Y, and Z, or captured for Specimen collection).
An Animal Source Record can be used directly for a Diagnostic (e.g., performing X-rays on a live animal at time t). The data model supports Diagnostics applied to the individual itself, rather than only to Specimens collected from the animal.
Live Animal Sources cannot be removed from the field (ex-situ). Only Carcasses of Animal Sources can be transported, stored, an necropsied in a facility.
An individual of species X recorded as part of a Group Source Record cannot also be included as an Animal Source Record, and vice versa. If an Event contains both a Group Source Record of species X and an Animal Source Record of species X, then the total number of animals of species X at the Event is the sum of the individuals recorded in the Group Source Record and the single individual recorded as an Animal Source Record.
For example, consider a herd of 20 cows illegally raised in a protected area, where their health is assessed at time t. If two cows are sampled individually, they are recorded as two separate Animal Source Record (one per sampled cow). The remaining 18 cows are documented as one Group Source Record, where the total number of individuals is categorized by sex, age, and health status. Thus, the total number of cows at the Event is 20 (18 from the Group Source Record + 2 from the two Group Source Records). To maintain the herd identity, a cluster can be used (see Complexities). If the Group Source Record initially included all 20 cows and two additional Animal Source Record were created for the sampled individuals, the total number of cows would incorrectly appear as 22—which must be avoided.
Vaccination
The data model allows the inclusion of vaccinations administered to an Animal Source at the time of capture or immobilization t. Multiple vaccination records can be added to an Animal Source Record as needed, ensuring that vaccination history accumulates for the corresponding Animal Source.
Carcass
Each Carcass recorded in the database originates from a single Animal Source and is collected at a specific time t. Therefore, a Carcass is always linked to one, and only one, Animal Source Record.
Key attributes of a Carcass include:
- Decomposition Condition
- Storage Ex-situ
- Storage during Transport
- Owner
- Availability
Just like Group Source carcasses, the carcass of an Animal Source may undergo through multiple shipments among different facilities and changes in storage over time. Both, the initial storage and changes in storage as well as shipments can be added cumulatively to the Animal Source, so the full history is available.
Carcasses do not provide Specimens directly; rather, Specimens are collected through the associated dead Animal Source Record. The origin of Specimens (primary necropsy, secondary necropsy, field, etc.) is tracked. Similarly, a Carcass is not directly used for Diagnostics. any diagnostic procedures that involve a full carcass, such as X-rays, applied to the corresponding dead Animal Source Record and documented accordingly.
Necropsy
A Necropsy is performed on a specific Animal Source Carcass. A Necropsy can take place is either at the time the dead animal is found (field necropsy) or after the Carcass has been collected in a facility.
Key attributes of a Carcass include:
- Necropsy Identifier
- Necropsy Cross Reference Identifier
- Necropsy Date
- Findings per system
- Availability
Necropsies can be classified as partial field necropsy, full necropsy, primary, and secondary. A primary necropsy typically begins with an intact Carcass that has not been previously examined in a facility. In contrast, a secondary necropsy is usually performed by a veterinary pathologist using either photographs from the primary Necropsy or conducting the Necropsy again.
A Necropsy does not provide Specimens directly; rather, Specimens are provided through the dead Animal Source Carcass.
The Necropsy findings are reported through text in fields such as “Gastrointestinal System”, “Reproductive System”, etc. The list of anatomical components encompassed by each of these elements or systems is provided in the table below and they follow the National Agricultural Library Thesaurus Concept Space (NALT), a controlled vocabulary of terms related to agricultural, biological, physical and social sciences (https://lod.nal.usda.gov/nalt/en/).
Mouth and Beak
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| teeth | https://lod.nal.usda.gov/nalt/32369 | dentine | |
| teeth | https://lod.nal.usda.gov/nalt/32369 | dentition | |
| beak | https://lod.nal.usda.gov/nalt/16134 | NA | |
| gingiva | https://lod.nal.usda.gov/nalt/9531 | Oral tissue surrounding and attached to teeth. | interdental papilla |
| gingiva | https://lod.nal.usda.gov/nalt/9531 | Oral tissue surrounding and attached to teeth. | gums (mouth) |
| lips | https://lod.nal.usda.gov/nalt/50308 | NA | |
| salivary glands | https://lod.nal.usda.gov/nalt/2899 | NA | |
| palate | https://lod.nal.usda.gov/nalt/26789 | roof of the mouth | |
| palate | https://lod.nal.usda.gov/nalt/26789 | hard palate | |
| palate | https://lod.nal.usda.gov/nalt/26789 | soft palate | |
| mouth | https://lod.nal.usda.gov/nalt/33560 | buccal cavity | |
| mouth | https://lod.nal.usda.gov/nalt/33560 | oral cavity | |
| mouth | https://lod.nal.usda.gov/nalt/33560 | periodontium | |
| tongue | https://lod.nal.usda.gov/nalt/53559 | NA |
Eyes
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| retina | https://lod.nal.usda.gov/nalt/38995 | The ten-layered nervous tissue membrane of the eye. It is continuous with the optic nerve and receives images of external objects and transmits visual impulses to the brain. Its outer surface is in contact with the choroid and the inner surface with the vitreous body. The outer-most layer is pigmented, whereas the inner nine layers are transparent. | NA |
| eye lens | https://lod.nal.usda.gov/nalt/38989 | NA | |
| sclera | https://lod.nal.usda.gov/nalt/290574 | The white, opaque, fibrous, outer tunic of the eyeball, covering it entirely excepting the segment covered anteriorly by the cornea. It is essentially avascular but contains apertures for vessels, lymphatics, and nerves. It receives the tendons of insertion of the extraocular muscles and at the corneoscleral junction contains the canal of Schlemm. | eye white |
| eyes | https://lod.nal.usda.gov/nalt/28392 | eye | |
| lacrimal apparatus | https://lod.nal.usda.gov/nalt/28564 | lacrimal gland | |
| lacrimal apparatus | https://lod.nal.usda.gov/nalt/28564 | conjunctival sac | |
| iris (eyes) | https://lod.nal.usda.gov/nalt/38993 | The most anterior portion of the uveal layer, separating the anterior chamber from the posterior. It consists of two layers - the stroma and the pigmented epithelium. Color of the iris depends on the amount of melanin in the stroma on reflection from the pigmented epithelium. | NA |
| conjunctiva | https://lod.nal.usda.gov/nalt/28561 | NA | |
| cornea | https://lod.nal.usda.gov/nalt/29206 | NA | |
| eyelids | https://lod.nal.usda.gov/nalt/38991 | NA |
Nose
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| nose | https://lod.nal.usda.gov/nalt/54351 | nostrils | |
| vomeronasal organ | https://lod.nal.usda.gov/nalt/308291 | An accessory chemoreceptor organ that is separated from the main olfactory mucosa. It is situated at the base of nasal septum close to the vomer and nasal bones. It forwards chemical signals (such as pheromones) to the central nervous system, thus influencing reproductive and social behavior. In humans, most of its structures except the vomeronasal duct undergo regression after birth. | Jacobson organ |
| vomeronasal organ | https://lod.nal.usda.gov/nalt/308291 | An accessory chemoreceptor organ that is separated from the main olfactory mucosa. It is situated at the base of nasal septum close to the vomer and nasal bones. It forwards chemical signals (such as pheromones) to the central nervous system, thus influencing reproductive and social behavior. In humans, most of its structures except the vomeronasal duct undergo regression after birth. | Jacobson's organ |
| vomeronasal organ | https://lod.nal.usda.gov/nalt/308291 | An accessory chemoreceptor organ that is separated from the main olfactory mucosa. It is situated at the base of nasal septum close to the vomer and nasal bones. It forwards chemical signals (such as pheromones) to the central nervous system, thus influencing reproductive and social behavior. In humans, most of its structures except the vomeronasal duct undergo regression after birth. | vomeronasal nerve |
| nasal mucosa | https://lod.nal.usda.gov/nalt/53591 | NA | |
| nasal cavity | https://lod.nal.usda.gov/nalt/54350 | NA |
Ears
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| inner ear | https://lod.nal.usda.gov/nalt/332326 | The essential part of the hearing organ consists of two labyrinthine compartments: the bony labyrinthine and the membranous labyrinth. The bony labyrinth is a complex of three interconnecting cavities or spaces (Cochlea; vestibular labyrinth; and semicircular canals) in the temporal bone. Within the bony labyrinth lies the membranous labyrinth which is a complex of sacs and tubules (cochlear duct; saccule and utricle; and semicircular ducts) forming a continuous space enclosed by epithelium and connective tissue. These spaces are filled with labyrinthine fluids of various compositions. | labyrinth (inner ear) |
| guttural pouch | https://lod.nal.usda.gov/nalt/35107 | NA | |
| ears | https://lod.nal.usda.gov/nalt/35074 | ear | |
| middle ear | https://lod.nal.usda.gov/nalt/332330 | The space and structures directly internal to the tympanic membrane and external to the inner ear (labyrinth). Its major components include the auditory ossicles and the eustachian tube that connects the cavity of middle ear (tympanic cavity) to the upper part of the throat. | tympanum |
| middle ear | https://lod.nal.usda.gov/nalt/332330 | The space and structures directly internal to the tympanic membrane and external to the inner ear (labyrinth). Its major components include the auditory ossicles and the eustachian tube that connects the cavity of middle ear (tympanic cavity) to the upper part of the throat. | tympanic cavity |
| otoliths | https://lod.nal.usda.gov/nalt/191711 | Structures of the inner ear which function in orientation and equilibrium sensing of vertebrates. Otoliths contain calcium concretions useful for species identification and age determination of fish, and provide information about their life history and ecology. | otoconia |
| otoliths | https://lod.nal.usda.gov/nalt/191711 | Structures of the inner ear which function in orientation and equilibrium sensing of vertebrates. Otoliths contain calcium concretions useful for species identification and age determination of fish, and provide information about their life history and ecology. | otolithic membrane |
| otoliths | https://lod.nal.usda.gov/nalt/191711 | Structures of the inner ear which function in orientation and equilibrium sensing of vertebrates. Otoliths contain calcium concretions useful for species identification and age determination of fish, and provide information about their life history and ecology. | statoconia |
| otoliths | https://lod.nal.usda.gov/nalt/191711 | Structures of the inner ear which function in orientation and equilibrium sensing of vertebrates. Otoliths contain calcium concretions useful for species identification and age determination of fish, and provide information about their life history and ecology. | otolites |
| ear canal | https://lod.nal.usda.gov/nalt/339897 | The narrow passage way that conducts the sound collected by the ear auricle to the tympanic membrane. | auditory canal |
| ear canal | https://lod.nal.usda.gov/nalt/339897 | The narrow passage way that conducts the sound collected by the ear auricle to the tympanic membrane. | meatus acusticus externus |
Sinuses
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Wounds and injuries | https://lod.nal.usda.gov/nalt/728 | NA | NA |
| Claws | https://lod.nal.usda.gov/nalt/26735 | NA | Talons |
| Claws | https://lod.nal.usda.gov/nalt/26735 | NA | Nails (non-primates) |
| Claws | https://lod.nal.usda.gov/nalt/26735 | NA | Claw horn |
| Skin (animal) | https://lod.nal.usda.gov/nalt/32499 | NA | NA |
| Fur | https://lod.nal.usda.gov/nalt/27376 | NA | Coat (animal) |
| Integument | https://lod.nal.usda.gov/nalt/9103 | NA | Exuviae |
| Integument | https://lod.nal.usda.gov/nalt/9103 | NA | Body wall |
| Hairs | https://lod.nal.usda.gov/nalt/44301 | NA | Scalp hair |
| Hairs | https://lod.nal.usda.gov/nalt/44301 | NA | Hairs (animal) |
| Antlers | https://lod.nal.usda.gov/nalt/10565 | NA | NA |
| Hooves | https://lod.nal.usda.gov/nalt/45820 | NA | Hoofs |
| Sebaceous glands | https://lod.nal.usda.gov/nalt/38913 | NA | Tarsal glands |
| Wattles | https://lod.nal.usda.gov/nalt/47472 | NA | NA |
| Scent glands | https://lod.nal.usda.gov/nalt/59050 | NA | Poll glands |
| Perineum | https://lod.nal.usda.gov/nalt/57466 | NA | NA |
| Sweat glands | https://lod.nal.usda.gov/nalt/38914 | NA | NA |
| Insect cuticle | https://lod.nal.usda.gov/nalt/9104 | NA | NA |
| Scales (integument) | https://lod.nal.usda.gov/nalt/47470 | NA | NA |
| Comb (integument) | https://lod.nal.usda.gov/nalt/28099 | NA | NA |
| Feathers | https://lod.nal.usda.gov/nalt/34442 | NA | NA |
| Nails (integument) | https://lod.nal.usda.gov/nalt/47469 | NA | Nails (primates) |
| Nails (integument) | https://lod.nal.usda.gov/nalt/47469 | NA | Nails (human) |
Integument
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Antlers | https://lod.nal.usda.gov/nalt/10565 | NA | NA |
| Claws | https://lod.nal.usda.gov/nalt/26735 | NA | Talons |
| Claws | https://lod.nal.usda.gov/nalt/26735 | NA | Nails (non-primates) |
| Claws | https://lod.nal.usda.gov/nalt/26735 | NA | Claw horn |
| Comb (integument) | https://lod.nal.usda.gov/nalt/28099 | NA | NA |
| Eyelashes | NA | NA | NA |
| Feathers | https://lod.nal.usda.gov/nalt/34442 | NA | NA |
| Fur | https://lod.nal.usda.gov/nalt/27376 | NA | Coat (animal) |
| Hairs | https://lod.nal.usda.gov/nalt/44301 | NA | Scalp hair |
| Hairs | https://lod.nal.usda.gov/nalt/44301 | NA | Hairs (animal) |
| Hooves | https://lod.nal.usda.gov/nalt/45820 | NA | Hoofs |
| Insect cuticle | https://lod.nal.usda.gov/nalt/9104 | NA | NA |
| Integument | https://lod.nal.usda.gov/nalt/9103 | NA | Exuviae |
| Integument | https://lod.nal.usda.gov/nalt/9103 | NA | Body wall |
| Nails (integument) | https://lod.nal.usda.gov/nalt/47469 | NA | Nails (primates) |
| Nails (integument) | https://lod.nal.usda.gov/nalt/47469 | NA | Nails (human) |
| Perineum | https://lod.nal.usda.gov/nalt/57466 | NA | NA |
| Scales (integument) | https://lod.nal.usda.gov/nalt/47470 | NA | NA |
| Scent glands | https://lod.nal.usda.gov/nalt/59050 | NA | Poll glands |
| Sebaceous glands | https://lod.nal.usda.gov/nalt/38913 | NA | Tarsal glands |
| Skin (animal) | https://lod.nal.usda.gov/nalt/32499 | NA | NA |
| Sweat glands | https://lod.nal.usda.gov/nalt/38914 | NA | NA |
| Wattles | https://lod.nal.usda.gov/nalt/47472 | NA | NA |
| Wounds and injuries | https://lod.nal.usda.gov/nalt/728 | NA | NA |
Skin
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Insect cuticle | https://lod.nal.usda.gov/nalt/9104 | NA | NA |
| Perineum | https://lod.nal.usda.gov/nalt/57466 | NA | NA |
| Scar | NA | NA | NA |
| Scent glands | https://lod.nal.usda.gov/nalt/59050 | NA | Poll glands |
| Sebaceous glands | https://lod.nal.usda.gov/nalt/38913 | NA | Tarsal glands |
| Skin (animal) | https://lod.nal.usda.gov/nalt/32499 | NA | NA |
| Sweat glands | https://lod.nal.usda.gov/nalt/38914 | NA | NA |
| Wounds and injuries | https://lod.nal.usda.gov/nalt/728 | NA | NA |
Subcutaneous Fat
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| subcutaneous fat | https://lod.nal.usda.gov/nalt/18338 | NA |
Muskuloskeletal System
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| joints (animal) | https://lod.nal.usda.gov/nalt/36135 | NA | |
| tendons | https://lod.nal.usda.gov/nalt/28572 | Fibrous bands or cords of connective tissue at the ends of muscle fibers that serve to attach the muscles to bones and other structures. | NA |
| bones | https://lod.nal.usda.gov/nalt/18497 | NA | |
| musculoskeletal system | https://lod.nal.usda.gov/nalt/8976 | skeletomuscular system | |
| musculoskeletal system | https://lod.nal.usda.gov/nalt/8976 | skeletal system | |
| skeletal muscle | https://lod.nal.usda.gov/nalt/35050 | NA | |
| fascia | https://lod.nal.usda.gov/nalt/28570 | NA | |
| skeleton | https://lod.nal.usda.gov/nalt/53793 | NA |
Body Cavities
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| mediastinum | https://lod.nal.usda.gov/nalt/51800 | NA | |
| body cavities | https://lod.nal.usda.gov/nalt/1610 | NA | |
| abdominal cavity | https://lod.nal.usda.gov/nalt/1609 | NA | |
| hemocoel | https://lod.nal.usda.gov/nalt/18328 | A body cavity of arthropods and some molluscs that contains blood or hemolymph, which functions as part of the circulatory system by directly bathing the organs. | haemocoel |
| thoracic cavity | https://lod.nal.usda.gov/nalt/18330 | NA |
Cardiovascular System
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Cardiovascular system | https://lod.nal.usda.gov/nalt/18060 | NA | NA |
| Heart | https://lod.nal.usda.gov/nalt/9341 | NA | Endocardium |
| Blood plasma | https://lod.nal.usda.gov/nalt/18062 | NA | NA |
| Arteries | https://lod.nal.usda.gov/nalt/10603 | The vessels carrying blood away from the heart. | NA |
| Inferior vena cava | https://lod.nal.usda.gov/nalt/190442 | The venous trunk which receives blood from the lower extremities and from the pelvic and abdominal organs. | NA |
| Blood veins | https://lod.nal.usda.gov/nalt/18176 | NA | NA |
| Blood vessels | https://lod.nal.usda.gov/nalt/12438 | NA | NA |
| Jugular vein | https://lod.nal.usda.gov/nalt/18177 | NA | NA |
| Coronary vessels | https://lod.nal.usda.gov/nalt/12440 | The veins and arteries of the heart. | Coronary arteries |
| Coronary sinus | https://lod.nal.usda.gov/nalt/190439 | A short vein that collects about two thirds of the venous blood from the myocardium and drains into the right atrium. Coronary sinus, normally located between the left atrium and left ventricle on the posterior surface of the heart, can serve as an anatomical reference for cardiac procedures. | NA |
| Hemolymph | https://lod.nal.usda.gov/nalt/26359 | Bloodlike fluid of the hemocoel in open circulatory systems of arthropods and most molluscs. | Haemolymph |
| Portal vein | https://lod.nal.usda.gov/nalt/18178 | NA | NA |
| Blood | https://lod.nal.usda.gov/nalt/18059 | NA | NA |
| Iliac artery | https://lod.nal.usda.gov/nalt/12442 | NA | NA |
| Hepatic artery | https://lod.nal.usda.gov/nalt/12441 | NA | NA |
| Superior vena cava | https://lod.nal.usda.gov/nalt/190443 | The venous trunk which returns blood from the head, neck, upper extremities and chest. | NA |
| Blood serum | https://lod.nal.usda.gov/nalt/18139 | The clear, watery portion of blood that separates out when blood coagulates. It lacks blood clotting factors such as fibrogen and prothrombin. | Serum |
| Carotid arteries | https://lod.nal.usda.gov/nalt/12439 | NA | NA |
| Caudal vein | https://lod.nal.usda.gov/nalt/254549 | NA | Tail vein |
| Myocardium | https://lod.nal.usda.gov/nalt/22397 | The muscle tissue of the heart. It is composed of striated, involuntary muscle cells connected to form the contractile pump to generate blood flow. | Cardiac muscle |
| Aorta | https://lod.nal.usda.gov/nalt/10602 | NA | Aortic root |
| Thymocytes | https://lod.nal.usda.gov/nalt/50929 | NA | NA |
| Fetal bovine serum | https://lod.nal.usda.gov/nalt/191461 | NA | Fetal calf serum |
| Fetal bovine serum | https://lod.nal.usda.gov/nalt/191461 | NA | Fbs (fetal bovine serum) |
| Pulmonary artery | https://lod.nal.usda.gov/nalt/12444 | NA | NA |
| Umbilical arteries | https://lod.nal.usda.gov/nalt/12445 | NA | NA |
| Saphenous vein | https://lod.nal.usda.gov/nalt/18179 | The vein which drains the foot and leg. | NA |
| Ductus arteriosus | https://lod.nal.usda.gov/nalt/28513 | NA | NA |
| Umbilical veins | https://lod.nal.usda.gov/nalt/18180 | NA | NA |
| Heart atrium | https://lod.nal.usda.gov/nalt/13743 | NA | NA |
| Vena cava | https://lod.nal.usda.gov/nalt/18181 | NA | Venae cavae |
| Heart valves | https://lod.nal.usda.gov/nalt/44757 | NA | NA |
| Heart ventricle | https://lod.nal.usda.gov/nalt/44758 | NA | NA |
| Mesenteric arteries | https://lod.nal.usda.gov/nalt/12443 | NA | NA |
Respiratory System
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Lungs | https://lod.nal.usda.gov/nalt/9345 | NA | NA |
| Larynx | https://lod.nal.usda.gov/nalt/49283 | NA | NA |
| Spiracles | https://lod.nal.usda.gov/nalt/61125 | NA | NA |
| Bronchi | https://lod.nal.usda.gov/nalt/20020 | The larger air passages of the lungs arising from the terminal bifurcation of the trachea. They include the largest two primary bronchi which branch out into secondary bronchi, and tertiary bronchi which extend into bronchioles and pulmonary alveoli. | Secondary bronchi |
| Bronchi | https://lod.nal.usda.gov/nalt/20020 | The larger air passages of the lungs arising from the terminal bifurcation of the trachea. They include the largest two primary bronchi which branch out into secondary bronchi, and tertiary bronchi which extend into bronchioles and pulmonary alveoli. | Primary bronchi |
| Bronchi | https://lod.nal.usda.gov/nalt/20020 | The larger air passages of the lungs arising from the terminal bifurcation of the trachea. They include the largest two primary bronchi which branch out into secondary bronchi, and tertiary bronchi which extend into bronchioles and pulmonary alveoli. | Bronchus |
| Bronchi | https://lod.nal.usda.gov/nalt/20020 | The larger air passages of the lungs arising from the terminal bifurcation of the trachea. They include the largest two primary bronchi which branch out into secondary bronchi, and tertiary bronchi which extend into bronchioles and pulmonary alveoli. | Tertiary bronchi |
| Air sacs | https://lod.nal.usda.gov/nalt/5852 | NA | NA |
| Vomeronasal organ | https://lod.nal.usda.gov/nalt/308291 | An accessory chemoreceptor organ that is separated from the main olfactory mucosa. It is situated at the base of nasal septum close to the vomer and nasal bones. It forwards chemical signals (such as pheromones) to the central nervous system, thus influencing reproductive and social behavior. In humans, most of its structures except the vomeronasal duct undergo regression after birth. | Jacobson organ |
| Vomeronasal organ | https://lod.nal.usda.gov/nalt/308291 | An accessory chemoreceptor organ that is separated from the main olfactory mucosa. It is situated at the base of nasal septum close to the vomer and nasal bones. It forwards chemical signals (such as pheromones) to the central nervous system, thus influencing reproductive and social behavior. In humans, most of its structures except the vomeronasal duct undergo regression after birth. | Jacobson's organ |
| Vomeronasal organ | https://lod.nal.usda.gov/nalt/308291 | An accessory chemoreceptor organ that is separated from the main olfactory mucosa. It is situated at the base of nasal septum close to the vomer and nasal bones. It forwards chemical signals (such as pheromones) to the central nervous system, thus influencing reproductive and social behavior. In humans, most of its structures except the vomeronasal duct undergo regression after birth. | Vomeronasal nerve |
| Trachea (vertebrates) | https://lod.nal.usda.gov/nalt/61127 | NA | NA |
| Bronchioles | https://lod.nal.usda.gov/nalt/266347 | The small airways branching off the tertiary bronchi. Terminal bronchioles lead into several orders of respiratory bronchioles which in turn lead into alveolar ducts and then into pulmonary alveoli. | NA |
| Respiratory system | https://lod.nal.usda.gov/nalt/5853 | NA | Respiratory tract |
| Respiratory mucosa | https://lod.nal.usda.gov/nalt/202662 | The mucous membrane lining the respiratory tract, including the nasal cavity; the larynx; the trachea; and the bronchi tree. The respiratory mucosa consists of various types of epithelial cells ranging from ciliated columnar to simple squamous, mucous goblet cells, and glands containing both mucous and serous cells. | Respiratory epithelium |
| Gills | https://lod.nal.usda.gov/nalt/9339 | NA | NA |
| Pleura | https://lod.nal.usda.gov/nalt/58808 | NA | NA |
| Tracheae (invertebrates) | https://lod.nal.usda.gov/nalt/61128 | NA | NA |
| Swim bladder | https://lod.nal.usda.gov/nalt/61126 | NA | Swimbladder |
| Swim bladder | https://lod.nal.usda.gov/nalt/61126 | NA | Gas bladder |
| Swim bladder | https://lod.nal.usda.gov/nalt/61126 | NA | Air bladder |
Gastrointestinal System
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Esophagus | https://lod.nal.usda.gov/nalt/33554 | NA | Oesophagus |
| Hepatopancreas | https://lod.nal.usda.gov/nalt/9342 | NA | NA |
| Intestines | https://lod.nal.usda.gov/nalt/42441 | NA | Intestinal tract |
| Nasopharynx | https://lod.nal.usda.gov/nalt/54355 | NA | Rhinopharynx |
| Nasopharynx | https://lod.nal.usda.gov/nalt/54355 | NA | Nasopharynges |
| Gastric mucosa | https://lod.nal.usda.gov/nalt/22390 | Lining of the stomach, consisting of an inner epithelium, a middle lamina propria, and an outer muscularis mucosae. The surface cells produce mucus that protects the stomach from attack by digestive acid and enzymes. When the epithelium invaginates into the lamina propria at various regions of the stomach, different tubular gastric glands are formed. These glands consist of cells that secrete mucus, enzymes, hydrochloric acid, or hormones. | Gastric glands |
| Gastric mucosa | https://lod.nal.usda.gov/nalt/22390 | Lining of the stomach, consisting of an inner epithelium, a middle lamina propria, and an outer muscularis mucosae. The surface cells produce mucus that protects the stomach from attack by digestive acid and enzymes. When the epithelium invaginates into the lamina propria at various regions of the stomach, different tubular gastric glands are formed. These glands consist of cells that secrete mucus, enzymes, hydrochloric acid, or hormones. | Cardiac glands |
| Gastric mucosa | https://lod.nal.usda.gov/nalt/22390 | Lining of the stomach, consisting of an inner epithelium, a middle lamina propria, and an outer muscularis mucosae. The surface cells produce mucus that protects the stomach from attack by digestive acid and enzymes. When the epithelium invaginates into the lamina propria at various regions of the stomach, different tubular gastric glands are formed. These glands consist of cells that secrete mucus, enzymes, hydrochloric acid, or hormones. | Pyloric glands |
| Gastric mucosa | https://lod.nal.usda.gov/nalt/22390 | Lining of the stomach, consisting of an inner epithelium, a middle lamina propria, and an outer muscularis mucosae. The surface cells produce mucus that protects the stomach from attack by digestive acid and enzymes. When the epithelium invaginates into the lamina propria at various regions of the stomach, different tubular gastric glands are formed. These glands consist of cells that secrete mucus, enzymes, hydrochloric acid, or hormones. | Stomach mucosa |
| Cloaca | https://lod.nal.usda.gov/nalt/26934 | NA | NA |
| Large intestine | https://lod.nal.usda.gov/nalt/23273 | NA | NA |
| Esophageal sphincter | https://lod.nal.usda.gov/nalt/38157 | NA | Oesophagogastric junction |
| Esophageal sphincter | https://lod.nal.usda.gov/nalt/38157 | NA | Esophagogastric junction |
| Esophageal sphincter | https://lod.nal.usda.gov/nalt/38157 | NA | Oesophageal sphincter |
| Liver | https://lod.nal.usda.gov/nalt/9344 | NA | Liver (anatomy) |
| Ileum | https://lod.nal.usda.gov/nalt/46899 | NA | NA |
| Colon | https://lod.nal.usda.gov/nalt/27880 | NA | NA |
| Rectum | https://lod.nal.usda.gov/nalt/10589 | NA | NA |
| Stomach | https://lod.nal.usda.gov/nalt/9350 | NA | NA |
| Crop (digestive system) | https://lod.nal.usda.gov/nalt/30108 | NA | Honey sac |
| Small intestine | https://lod.nal.usda.gov/nalt/34888 | NA | NA |
| Rumen | https://lod.nal.usda.gov/nalt/41231 | NA | NA |
| Pancreas | https://lod.nal.usda.gov/nalt/2897 | NA | NA |
| Anus | https://lod.nal.usda.gov/nalt/8441 | NA | Anal sphincter |
| Cecum | https://lod.nal.usda.gov/nalt/20837 | NA | Caecum |
| Gall bladder | https://lod.nal.usda.gov/nalt/9338 | NA | Gallbladder |
| Omasum | https://lod.nal.usda.gov/nalt/41229 | NA | NA |
| Duodenum | https://lod.nal.usda.gov/nalt/34887 | NA | NA |
| Gizzard | https://lod.nal.usda.gov/nalt/33557 | NA | NA |
| Gastric fundus | https://lod.nal.usda.gov/nalt/42409 | NA | Stomach fundus |
| Gastrointestinal system | https://lod.nal.usda.gov/nalt/33556 | NA | NA |
| Reticulum | https://lod.nal.usda.gov/nalt/41230 | NA | NA |
| Pharynx | https://lod.nal.usda.gov/nalt/33561 | NA | Pharynges |
| Oropharynx | https://lod.nal.usda.gov/nalt/234524 | The middle portion of the pharynx that lies posterior to the mouth, inferior to the soft palate, and superior to the base of the tongue and epiglottis. It has a digestive function as food passes from the mouth into the oropharynx before entering esophagus. | NA |
| Abomasum | https://lod.nal.usda.gov/nalt/1770 | NA | NA |
| Jejunum | https://lod.nal.usda.gov/nalt/48097 | NA | NA |
| Pylorus | https://lod.nal.usda.gov/nalt/60608 | NA | NA |
| Proventriculus | https://lod.nal.usda.gov/nalt/33562 | NA | NA |
Urinary Tract
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Ureter | https://lod.nal.usda.gov/nalt/66771 | NA | NA |
| Urethra | https://lod.nal.usda.gov/nalt/66772 | NA | NA |
| Kidneys | https://lod.nal.usda.gov/nalt/9343 | NA | Head kidney |
| Kidneys | https://lod.nal.usda.gov/nalt/9343 | NA | Anterior kidney |
| Bladder | https://lod.nal.usda.gov/nalt/17878 | A musculomembranous sac along the urinary tract. Urine flows from the kidneys into the bladder via the ureters (ureter), and is held there until urination. | Urinary bladder |
| Urinary tract | https://lod.nal.usda.gov/nalt/17881 | NA | NA |
Reproductive System
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Spermatic cord | https://lod.nal.usda.gov/nalt/51286 | NA | NA |
| Animal ovaries | https://lod.nal.usda.gov/nalt/29278 | NA | Mammalian ovary |
| Female reproductive system | https://lod.nal.usda.gov/nalt/24114 | NA | NA |
| Oviductal ampulla | https://lod.nal.usda.gov/nalt/8132 | NA | NA |
| Cervix | https://lod.nal.usda.gov/nalt/24102 | NA | Cervix uteri |
| Cervix | https://lod.nal.usda.gov/nalt/24102 | NA | Uterine cervix |
| Male reproductive system | https://lod.nal.usda.gov/nalt/20336 | NA | Gubernaculum |
| Cloaca | https://lod.nal.usda.gov/nalt/26934 | NA | NA |
| Epididymis | https://lod.nal.usda.gov/nalt/22094 | NA | NA |
| Caput epididymidis | https://lod.nal.usda.gov/nalt/22093 | NA | NA |
| Gynoecium | https://lod.nal.usda.gov/nalt/22619 | A collective term for the female reproductive structures of a flower. | NA |
| Ovipositor | https://lod.nal.usda.gov/nalt/39793 | NA | NA |
| Uterine tissue | https://lod.nal.usda.gov/nalt/9538 | NA | NA |
| Uterus | https://lod.nal.usda.gov/nalt/39795 | NA | NA |
| Seminiferous tubules | https://lod.nal.usda.gov/nalt/62834 | NA | NA |
| Prostate gland | https://lod.nal.usda.gov/nalt/38912 | NA | NA |
| Oviducts | https://lod.nal.usda.gov/nalt/39792 | Ducts that serve exclusively for the passage of eggs from the ovaries to the exterior of the body. In non-mammals, they are termed oviducts. In mammals, they are highly specialized and known as fallopian tubes. | NA |
| Rete testis | https://lod.nal.usda.gov/nalt/61147 | NA | NA |
| Prepuce | https://lod.nal.usda.gov/nalt/57285 | NA | NA |
| Shell gland | https://lod.nal.usda.gov/nalt/9347 | The specialized glandular part of the oviduct that forms the egg's shell. | Egg shell gland |
| Shell gland | https://lod.nal.usda.gov/nalt/9347 | The specialized glandular part of the oviduct that forms the egg's shell. | Eggshell gland |
| Scrotum | https://lod.nal.usda.gov/nalt/51285 | NA | NA |
| Cervical mucus | https://lod.nal.usda.gov/nalt/24105 | NA | NA |
| Myometrium | https://lod.nal.usda.gov/nalt/54164 | NA | NA |
| Bulbourethral gland | https://lod.nal.usda.gov/nalt/20331 | NA | Bulbo-urethral gland |
| Oviductal isthmus | https://lod.nal.usda.gov/nalt/47917 | NA | NA |
| Ovarian follicles | https://lod.nal.usda.gov/nalt/30722 | NA | Subordinate follicles |
| Ovarian follicles | https://lod.nal.usda.gov/nalt/30722 | NA | Dominant follicles |
| Vaginal mucosa | https://lod.nal.usda.gov/nalt/53592 | NA | NA |
| Testes | https://lod.nal.usda.gov/nalt/43422 | NA | Testicles |
| Testes | https://lod.nal.usda.gov/nalt/43422 | NA | Testis |
| Embryo sac | https://lod.nal.usda.gov/nalt/36821 | NA | NA |
| Accessory sex glands | https://lod.nal.usda.gov/nalt/2139 | NA | Accessory sex organs |
| Accessory sex glands | https://lod.nal.usda.gov/nalt/2139 | NA | Male ampulla |
| Accessory sex glands | https://lod.nal.usda.gov/nalt/2139 | NA | Ampulla (male) |
| Reproductive system | https://lod.nal.usda.gov/nalt/2141 | NA | NA |
| Inferior ovary | https://lod.nal.usda.gov/nalt/333770 | An inferior ovary is one that is situated entirely below the point of attachment of the other floral parts in a flower. | NA |
| Endometrium | https://lod.nal.usda.gov/nalt/37037 | NA | NA |
| Cauda epididymidis | https://lod.nal.usda.gov/nalt/23129 | NA | NA |
| Ductus deferens | https://lod.nal.usda.gov/nalt/34844 | The excretory duct of the testes that carries spermatozoa. It rises from the scrotum and joins the seminal vesicles to form the ejaculatory duct. | NA |
| Penis | https://lod.nal.usda.gov/nalt/51284 | NA | NA |
| Fallopian tubes | https://lod.nal.usda.gov/nalt/188598 | A pair of highly specialized muscular canals extending from the uterus to its corresponding ovary. They provide the means for ovum collection, and the site for the final maturation of gametes and fertilization. | Uterine tubes |
| Fallopian tubes | https://lod.nal.usda.gov/nalt/188598 | A pair of highly specialized muscular canals extending from the uterus to its corresponding ovary. They provide the means for ovum collection, and the site for the final maturation of gametes and fertilization. | Salpinges |
| Fallopian tubes | https://lod.nal.usda.gov/nalt/188598 | A pair of highly specialized muscular canals extending from the uterus to its corresponding ovary. They provide the means for ovum collection, and the site for the final maturation of gametes and fertilization. | Mammalian oviducts |
| Superior ovary | https://lod.nal.usda.gov/nalt/333769 | A superior ovary is one that is situated entirely above the point of attachment of the other floral parts in a flower. | NA |
| Vagina | https://lod.nal.usda.gov/nalt/39796 | NA | NA |
| Seminal vesicles | https://lod.nal.usda.gov/nalt/2142 | NA | Vesicular gland |
| Corpus epididymidis | https://lod.nal.usda.gov/nalt/29276 | NA | NA |
| Vulva | https://lod.nal.usda.gov/nalt/39797 | NA | NA |
Lymphatic System
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Spleen | https://lod.nal.usda.gov/nalt/9348 | NA | NA |
| Peyer's patches | https://lod.nal.usda.gov/nalt/50921 | Lymphoid tissue on the mucosa of the small intestine. | Peyer patches |
| Adenoids | https://lod.nal.usda.gov/nalt/240180 | A collection of lymphoid nodules on the posterior wall and roof of the nasopharynx. | Pharyngeal tonsils |
| Bursa of fabricius | https://lod.nal.usda.gov/nalt/20540 | NA | Bursa fabricii |
| Lymph nodes | https://lod.nal.usda.gov/nalt/50915 | NA | Lymphnodes |
| Lymphatic system | https://lod.nal.usda.gov/nalt/20541 | NA | NA |
| Thymus gland | https://lod.nal.usda.gov/nalt/37012 | NA | NA |
| Tonsils | https://lod.nal.usda.gov/nalt/9351 | A round-to-oval mass of lymphoid tissue embedded in the lateral wall of the pharynx. There is one on each side of the oropharynx in the fauces between the anterior and posterior pillars of the soft palate. | Palatine tonsils |
| Lymph | https://lod.nal.usda.gov/nalt/18343 | NA | NA |
| Thoracic duct | https://lod.nal.usda.gov/nalt/50922 | NA | NA |
Endocrine System
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Posterior pituitary | https://lod.nal.usda.gov/nalt/54725 | Neural tissue of the pituitary gland, also known as the neurohypophysis. It consists of the distal axons of neurons that produce vasopressin and oxytocinin in the supraoptic nucleus and the paraventricular nucleus. These axons travel down through the median eminence, the hypothalamic infundibulum of the pituitary stalk, to the posterior lobe of the pituitary gland. | Neurohypophysis |
| Posterior pituitary | https://lod.nal.usda.gov/nalt/54725 | Neural tissue of the pituitary gland, also known as the neurohypophysis. It consists of the distal axons of neurons that produce vasopressin and oxytocinin in the supraoptic nucleus and the paraventricular nucleus. These axons travel down through the median eminence, the hypothalamic infundibulum of the pituitary stalk, to the posterior lobe of the pituitary gland. | Posterior pituitary gland |
| Posterior pituitary | https://lod.nal.usda.gov/nalt/54725 | Neural tissue of the pituitary gland, also known as the neurohypophysis. It consists of the distal axons of neurons that produce vasopressin and oxytocinin in the supraoptic nucleus and the paraventricular nucleus. These axons travel down through the median eminence, the hypothalamic infundibulum of the pituitary stalk, to the posterior lobe of the pituitary gland. | Infundibular stalk |
| Thyroid gland | https://lod.nal.usda.gov/nalt/37013 | NA | Thyroid (gland) |
| Parathyroid glands | https://lod.nal.usda.gov/nalt/37009 | NA | NA |
| Pituitary gland | https://lod.nal.usda.gov/nalt/9900 | A small, unpaired gland situated in the sella turcica tissue. It is connected to the hypothalamus by a short stalk. | Hypophysis |
| Endocrine system | https://lod.nal.usda.gov/nalt/8971 | NA | NA |
| Adrenal cortex | https://lod.nal.usda.gov/nalt/4339 | NA | Zona fasciculata |
| Adrenal cortex | https://lod.nal.usda.gov/nalt/4339 | NA | Zona reticularis |
| Adrenal cortex | https://lod.nal.usda.gov/nalt/4339 | NA | Zona glomerulosa |
| Thymus gland | https://lod.nal.usda.gov/nalt/37012 | NA | NA |
| Endocrine glands | https://lod.nal.usda.gov/nalt/4351 | Ductless glands that secrete hormones directly into the blood circulation. These hormones influence the metabolism and other functions of cells in the body. | NA |
| Adrenal glands | https://lod.nal.usda.gov/nalt/4343 | NA | NA |
| Adrenal medulla | https://lod.nal.usda.gov/nalt/4352 | NA | NA |
| Anterior pituitary | https://lod.nal.usda.gov/nalt/4132 | The anterior glandular lobe of the pituitary gland, also known as the adenohypophysis. It secretes the adenohypophyseal hormones that regulate vital functions such as growth; metabolism; and reproduction. | Adenohypophysis |
| Anterior pituitary | https://lod.nal.usda.gov/nalt/4132 | The anterior glandular lobe of the pituitary gland, also known as the adenohypophysis. It secretes the adenohypophyseal hormones that regulate vital functions such as growth; metabolism; and reproduction. | Anterior pituitary gland |
Nervous System
| Topography | NALT_URI | Definition_NALT | Alternative_name |
|---|---|---|---|
| Ganglia | https://lod.nal.usda.gov/nalt/1617 | NA | Ganglion |
| Cerebrospinal fluid | https://lod.nal.usda.gov/nalt/18339 | NA | NA |
| Optic lobe | https://lod.nal.usda.gov/nalt/19228 | NA | NA |
| Meninges | https://lod.nal.usda.gov/nalt/23678 | NA | NA |
| Mesencephalon | https://lod.nal.usda.gov/nalt/334166 | The middle of the three primitive cerebral vesicles of the embryonic brain. without further subdivision, midbrain develops into a short, constricted portion connecting the pons and the diencephalon. midbrain contains two major parts, the dorsal tectum mesencephali and the ventral tegmentum mesencephali, housing components of auditory, visual, and other sensorimoter systems. | NA |
| Optic tectum | https://lod.nal.usda.gov/nalt/334171 | The anterior pair of the quadrigeminal bodies which coordinate the general behavioral orienting responses to visual stimuli, such as whole-body turning, and reaching. | Anterior colliculus |
| Optic tectum | https://lod.nal.usda.gov/nalt/334171 | The anterior pair of the quadrigeminal bodies which coordinate the general behavioral orienting responses to visual stimuli, such as whole-body turning, and reaching. | Superior colliculi |
| Optic tectum | https://lod.nal.usda.gov/nalt/334171 | The anterior pair of the quadrigeminal bodies which coordinate the general behavioral orienting responses to visual stimuli, such as whole-body turning, and reaching. | Superior colliculus |
| Hippocampus | https://lod.nal.usda.gov/nalt/19223 | A curved elevation of gray matter extending the entire length of the floor of the temporal horn of the lateral ventricle. | Ammon's horn |
| Hippocampus | https://lod.nal.usda.gov/nalt/19223 | A curved elevation of gray matter extending the entire length of the floor of the temporal horn of the lateral ventricle. | Hippocampal formation |
| Amygdala | https://lod.nal.usda.gov/nalt/8210 | Almond-shaped group of basal nuclei anterior to the inferior horn of the lateral ventricle of the temporal lobe. The amygdala is part of the limbic system. | Archistriatum |
| Amygdala | https://lod.nal.usda.gov/nalt/8210 | Almond-shaped group of basal nuclei anterior to the inferior horn of the lateral ventricle of the temporal lobe. The amygdala is part of the limbic system. | Massa intercalata |
| Amygdala | https://lod.nal.usda.gov/nalt/8210 | Almond-shaped group of basal nuclei anterior to the inferior horn of the lateral ventricle of the temporal lobe. The amygdala is part of the limbic system. | Amygdaloid nuclear complex |
| Amygdala | https://lod.nal.usda.gov/nalt/8210 | Almond-shaped group of basal nuclei anterior to the inferior horn of the lateral ventricle of the temporal lobe. The amygdala is part of the limbic system. | Corpus amygdaloideum |
| Amygdala | https://lod.nal.usda.gov/nalt/8210 | Almond-shaped group of basal nuclei anterior to the inferior horn of the lateral ventricle of the temporal lobe. The amygdala is part of the limbic system. | Intercalated amygdaloid nuclei |
| Amygdala | https://lod.nal.usda.gov/nalt/8210 | Almond-shaped group of basal nuclei anterior to the inferior horn of the lateral ventricle of the temporal lobe. The amygdala is part of the limbic system. | Amygdaloid body |
| Amygdala | https://lod.nal.usda.gov/nalt/8210 | Almond-shaped group of basal nuclei anterior to the inferior horn of the lateral ventricle of the temporal lobe. The amygdala is part of the limbic system. | Nucleus amygdalae |
| Spinal cord | https://lod.nal.usda.gov/nalt/23679 | NA | NA |
| Cerebellum | https://lod.nal.usda.gov/nalt/8211 | NA | NA |
| Cerebral cortex | https://lod.nal.usda.gov/nalt/24050 | NA | NA |
| Thalamus | https://lod.nal.usda.gov/nalt/19229 | Paired bodies containing mostly gray substance and forming part of the lateral wall of the third ventricle of the brain. The thalamus represents the major portion of the diencephalon and is commonly divided into cellular aggregates known as nuclear groups. | NA |
| Brain | https://lod.nal.usda.gov/nalt/9337 | NA | Forebrain |
| Brain | https://lod.nal.usda.gov/nalt/9337 | NA | Midbrain |
| Brain | https://lod.nal.usda.gov/nalt/9337 | NA | Protocerebrum |
| Brain | https://lod.nal.usda.gov/nalt/9337 | NA | Tritocerebrum |
| Brain | https://lod.nal.usda.gov/nalt/9337 | NA | Deutocerebrum |
| Brain | https://lod.nal.usda.gov/nalt/9337 | NA | Hindbrain |
| Thoracic ganglia | https://lod.nal.usda.gov/nalt/42317 | NA | Thoracic ganglion |
| Ependyma | https://lod.nal.usda.gov/nalt/79494 | A thin membrane that lines the ventricles of the brain and the central canal of the spinal cord. | NA |
| Hypothalamus | https://lod.nal.usda.gov/nalt/19224 | NA | NA |
| Brain stem | https://lod.nal.usda.gov/nalt/19221 | The part of the brain that connects the cerebral hemispheres with the spinal cord. It consists of the mesencephalon; pons; and medulla oblongata. | Locus coeruleus |
| Brain stem | https://lod.nal.usda.gov/nalt/19221 | The part of the brain that connects the cerebral hemispheres with the spinal cord. It consists of the mesencephalon; pons; and medulla oblongata. | Brainstem |
| Brain stem | https://lod.nal.usda.gov/nalt/19221 | The part of the brain that connects the cerebral hemispheres with the spinal cord. It consists of the mesencephalon; pons; and medulla oblongata. | Truncus cerebri |
| Medulla oblongata | https://lod.nal.usda.gov/nalt/19225 | NA | NA |
| Central nervous system | https://lod.nal.usda.gov/nalt/19220 | NA | NA |
| Neocortex | https://lod.nal.usda.gov/nalt/199435 | NA | NA |
| Abdominal ganglia | https://lod.nal.usda.gov/nalt/1615 | NA | Abdominal ganglion |
| Subesophageal ganglia | https://lod.nal.usda.gov/nalt/42316 | NA | NA |
| Cerebrum | https://lod.nal.usda.gov/nalt/19222 | NA | NA |
| Frontal lobe | https://lod.nal.usda.gov/nalt/286312 | NA | Frontal cortex |
| Motor cortex | https://lod.nal.usda.gov/nalt/281208 | NA | NA |
| Preoptic area | https://lod.nal.usda.gov/nalt/46761 | NA | NA |
| Paraventricular hypothalamic nucleus | https://lod.nal.usda.gov/nalt/46760 | NA | Paraventricular nucleus of the hypothalamus |
| Cerebral ventricles | https://lod.nal.usda.gov/nalt/24054 | NA | NA |
| Prefrontal cortex | https://lod.nal.usda.gov/nalt/286315 | NA | Orbital cortex |
| Prefrontal cortex | https://lod.nal.usda.gov/nalt/286315 | NA | Orbitofrontal cortex |
| Olfactory bulb | https://lod.nal.usda.gov/nalt/19227 | NA | NA |
Environmental Source
An Environmental Source is a unit in space where Specimens that cannot be associated with a Group Source, an Animal Source, or Arthropod Source can be collected from (e.g., the site where feces of an unknown animal are found).
Key attributes of a Environmental Source include:
- Environmental Source ID
- Environmental Source Cross Reference ID
- Environment Type (e.g., pond, grassland, landfill)
Environmental Source Record
An Environmental Source Record is the site where environmental Specimens are collected from at time t. Many types of abiotic or biotic tissue can be obtained from the same Environmental Source Record, for example, a specific site in a pond at time t where water, sediment, and air are obtained. Moreover, Environmental Source Record can contain Diagnostics, for example, the turbidity of the water at the site.
Key attributes of a Environmental Source Record include:
- Environmental Source Record ID
- Record Number
If multiple tissue types are obtained a time t from an Environmental Source and each of them is associated with a particular active collection effort, then there will be multiple Collection Points under the same Event containing the same Environmental Source. In this case it is key to properly enter the Environmental Source Record IDs.
Arthropod Source
An Arthropod Source is a unit in space where arthropods exist and they can be taken from (e.g., a household in the forest where traps can be set).
Key attributes of a Arthropod Source include:
- Source ID
- Cross Reference ID
- Cross Reference ID Origin
If the interest of the Surveillance Activity is at the arthropod individual level (e.g., butterflies with problems in their wings or with parasites), then the user should consider these arthropods as Animal Sources. Arthropods from Animals Sources (attached ticks, lice, fleas, mites) are Specimens (see next section) from those Animal Source and not Arthropod Sources.
Arthropod Source Record
An Arthropod Source Record represents the set of arthropods collected from an Arthropod Source at time t using a specific method. For example, the set of mosquitoes at time t (Arthropod Source Record) from a site in the forest (Arthropod Source) captured using a CO₂ trap (Collection Point). This means that mosquitoes obtained using CO₂ traps and BG traps (Collection Points) at an Arthropod Source at time t belong to the same Arthropod Source Record.
Key attributes of a Arthropod Source Record include:
- Arthropod Source Record Number
- Arthropod Species
- Number by age, sex, and condition (females only)
Similar to Environmental Sources, a key distinction between Arthropod Sources and Group or Animal Sources is that an Event can contain multiple Collection Points (e.g., each trap) containing the same Arthropod Record Record.
A key characteristic of an Arthropods Source Record is that “species” is a property of the Source for Group and Animal Sources, while for Arthropod Sources, there can be several species under the Source Record level. For example, mosquitoes collected from a specific site (Source) using CO₂ traps (Collection Point) will be identified and counted by species in the Arthropod Source Record.
If an arthropod Collection attempt at time t fails, the Collection itself is recorded, and an empty Arthropod Source Record is added.
Specimen
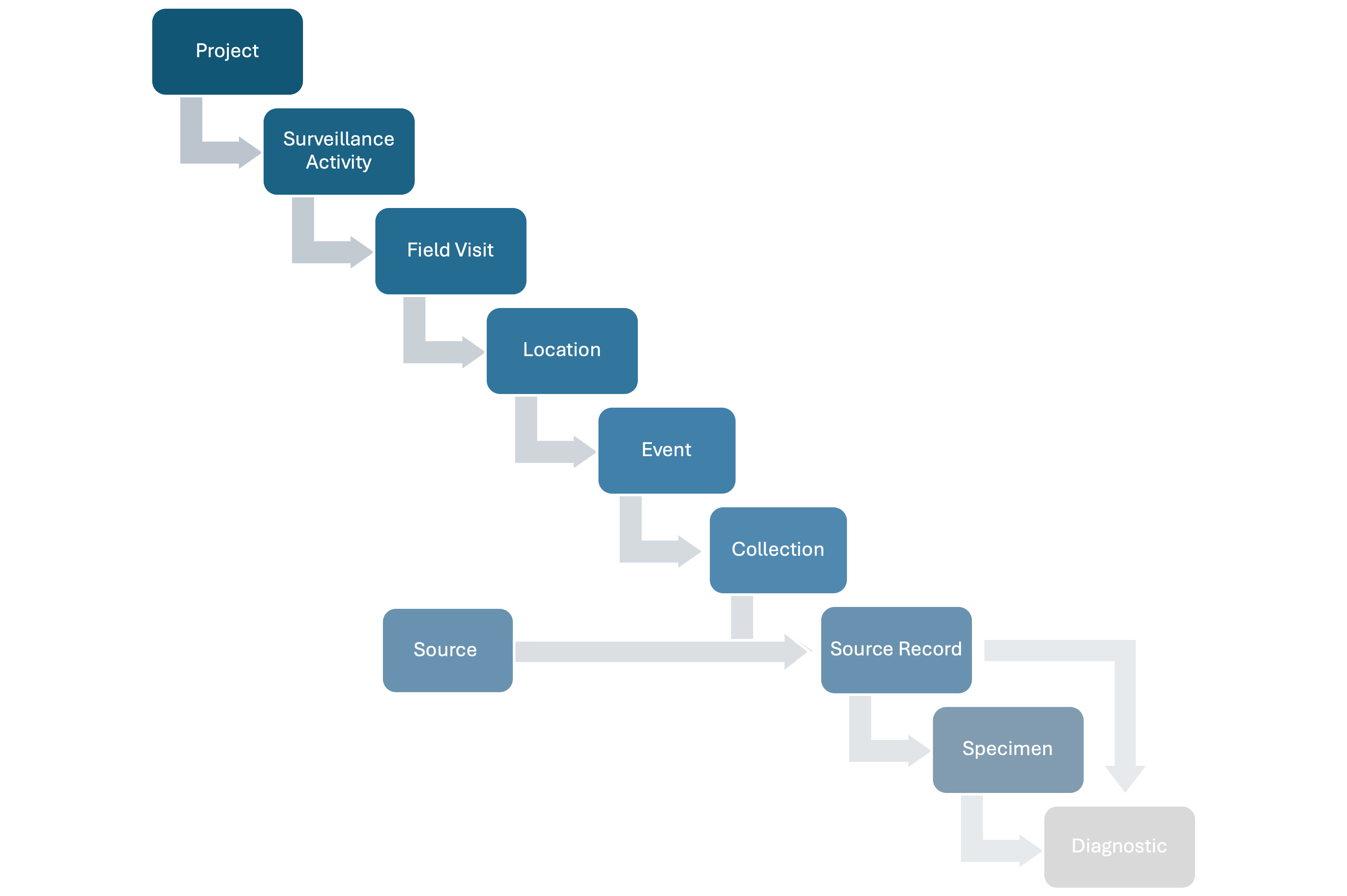
Generalities
In the data model, Specimens refer to tissues collected to conduct Diagnostics (see Diagnostics).
Specimens originate from the following:
Sampled Group and Animal Source Records (i.e., an oral swab from an individual animal). These Specimens may consist of either a single type of tissue (e.g., blood) or multiple tissue types (e.g., mixed blood and saliva) from the same Source Record.
Pools of arthropods built from the arthropods present at an Arthropod Source Record.
A Diagnostic Product created by running a Diagnostic on a Specimen that can be used in further Diagnostics (i.e., cDNA created from RNA in a sample)
Other Specimens (Pooled Specimen). For example, different Specimens from the same or multiple Source Records are mixed (grey boxes in figure below) with Diagnostic Products. In HAWK, a pooled specimen is built from existing Specimens records.
Key attributes of a Specimen include:
- Specimen ID
- Tissue Type
- Specimen Original Amount
- Specimen Current Quantity Stored
- Origin
- Ownership
Specimens are stored, and their storage may change multiple times as they are transferred within and between facilities or because the amount of Specimen changes as it is used in a Diagnostic. Similarly, Specimens can be shipped multiple times.
Specimens without any amount of tissue left remain in HAWK so the last storage, shipment, and use in Diagnostics can be traced.
Specimens of Group Source Records
An example of Group Source Specimen are feces collected from the bottom of a cage with animals of the same species but it is unknown which of the animals dropped the feces.
Group Source Specimens cannot be obtained AFTER the last date the Group Source was observed (the last Record of the Group Source). For example, it is possible to collect feces from a cage that restrains a Group Source longitudinally and also from the same cage after the animals of the Group Source were moved. In this case, the Specimen from the empty cage does not belong to the Group Source Record but to an Environmental Source Record. The sampling of the feces from the empty cage belongs to a different Event. To keep the connection between the corresponding Specimens, it is possible to cluster the Group Source Records and the Environmental Source Record (see Complexities).
Specimens of Animal Source Records
An example of Animal Source Specimen is 2 ml of blood taken from a lion.
Animal Source Specimens cannot be obtained AFTER the last date the Animal Source was observed (the last Record of the Animal Source). For example, it is possible to collect feces from a cage that restrained an Animal Source longitudinally and also from the same cage after the animal was moved. In this case, the Specimen from the empty cage does not belong to the Animal Source Record but to an Environmental Source Record. The sampling of the feces from the empty cage belongs to a different Event. To keep the connection between the corresponding Specimens, it is possible to cluster the Animal Source Records and the Environmental Source Record (see Complexities).
The data model DOES accept Animal Source Specimens that are CREATED after the last date the Animal Source was documented (the last Record of the Animal Source). New Specimens can be generated during a Necropsy or from a stored Carcass. In this case, the date of Specimen creation is not necessarily the date when the Animal Source was found, when the carcass was collected, or when the animal died, but after the storage of the carcass or the date of the Necropsy. It is possible to track if a Specimen was collected in the field (from the animal, carcass, during a field necropsy, or the ground near the animal), or in a facility from the carcass or during a primary or secondary Necropsy based on the information entered for Specimens.
Specimens from Environmental Source Records
An example of Environmental Source Specimen is 10 grams of soil collected from a landfill (Environmental Source). Another useful example are feces of unknown origin found in the field at time t (Source Record) at a specific site.
The data model DOES accept Arthropod Source Specimens that were CREATED after the last date the Arthropod Source was visited (the last Record of the Environmental Source).
Specimens from Arthropod Source Records
An example of Arthropod Source Specimen is a mosquito pool created from the set of mosquitoes captured at the “Arthropod Source** site at time t.
The data model DOES accept Arthropod Source Specimens that were CREATED after the last date the Arthropod Source was visited (the last Record of the Environmental Source).
Specimens with a mix of tissues coming from a unique or several Source Records
A bat is swabbed in its oral cavity and in its rectum but then the swabs are placed together in a tube and considered a single Specimen. Then, there is a single Specimen with a unique Source Record origin and two types of tissue: “rectal swab” and “oral swab” in a tube.
It is possible also to get blood from two bats and mix it. The data model can accommodate this case because Specimens can have multiple Source Record origins. In the example, both bled bats (Source Records) are the origin of a single Specimen with “blood” tissue consisting in mm of blood.
Similarly, it is possible to generate a Specimen by mixing tissue of different type from different Source Records. For example, the blood (tissue) of a bat captured at time t (Animal Source Record) with the feces (tissue) collected from the bottom of the roost of the same bat at time t (Group Source Record). In this case, the mixed tissue (blood from an Animal Source and Feces from a Group Source) can be included in the data model as a single Specimen with two Source Records as origin and two types of tissue.
The key here is that Specimens are generated by mixing tissue coming from different Source Records, which is different than mixing Specimens. In the data model, mixing Specimens means mixing data Units already documented (see “Pooled Specimens” below).
Specimens from Diagnostic Products
Products generated by a Diagnostic method (see Diagnostic Products as Specimens below) can be stored and used as Specimen in further Diagnostics (e.g., use cDNA created as part of Diagnostic A used in a new RT-PCR, Diagnostic B). The data model can accommodate Diagnostic Products to be used as Specimens in future Diagnostics. In this case, the origin of these new “Specimens” are specific Diagnostics and not Source Records, and their type is a diagnostic product such as cDNA. The remaining properties of a Specimen from a Diagnostic Product are the same as for Specimens from Source Records (see Data Dictionary).
Diagnostic Products must be added to the data model as a Specimen, so it can have Diagnostics and be pooled with other Specimens.
Pooled Specimens
Specimens can be created by mixing other Specimens of any origin. For example, mixing a Specimen from an Arthropod Source, a Specimen from an Animal Source, a Specimen from an Environmental Source, and a Specimen from an Arthropod Source (see below). The origin of Pooled Specimens is tracked in the data model. In the example, the origin of the Specimen is four Specimens and the new Specimen has potentially four types of tissue. The remaining properties are the same as for Specimens from Source Records (see Data Dictionary).
Specimens in Containers
In situations where space or materials are limited, it is possible that multiple Specimens are stored in a unique container. This approach is clearly not ideal because it can lead to cross-contamination and make actual Specimen tracing more complex. However, the data model has a Container Identifier as a Specimen property (see Data Dictionary). Specific properties for each individual Specimen within the container, such as type, quantity, etc, should allow their visual identification within the container.
Diagnostics
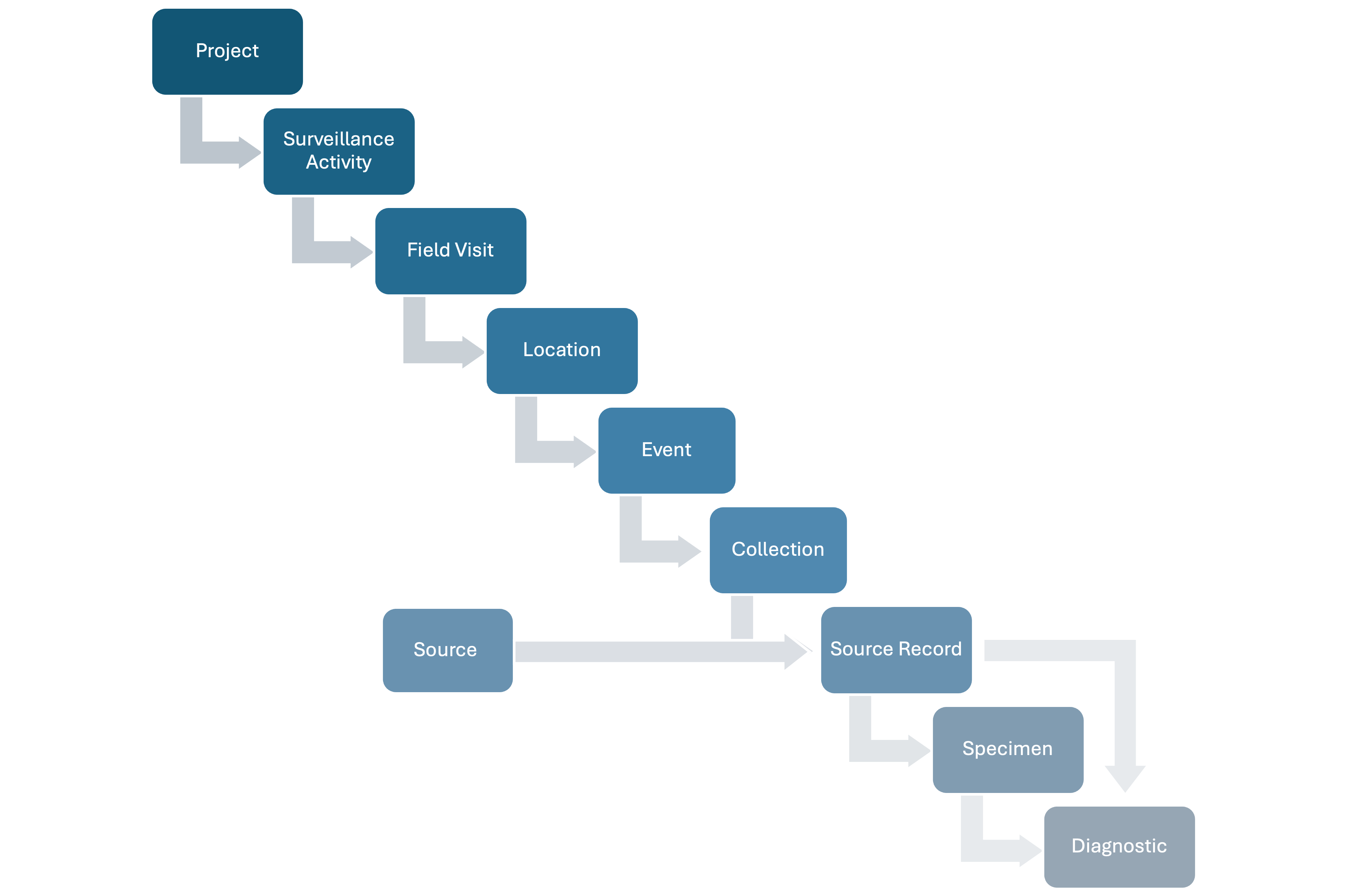
Diagnostics in the data model encompass various objective techniques used to identify hazards (biological, chemical, physical) or physiological problems in either Records of Animal, Group, and Environmental Sources or Specimens obtained from Source Records of any type. Diagnostics conducted directly in Group Animal, and Environmental Sources can include general body condition (Group or Animal Source), the width of body fat (Group or Animal Source), an ultrasound (Animal Source), and turbidity of water (Environmental Source). Diagnostics can range from advanced techniques such as metagenomics to standardize observations of body condition. Whatever the Diagnostic methods used are, they must be reported in the metadata of the Surveillance Activity.
In the data model, Diagnostics can be conducted in a Laboratory or similar (see Laboratory below) but it is also possible to include field-based assays. Diagnostics can also be conducted by an individual (ranger, hunter, researcher, biologist, etc.) when they involve external or simple observations such as the body condition of an animal to assess a nutrition related hazard.
In the data model, a specific Diagnostic can target single or multiple hazards. Therefore, each Diagnostic can have multiple results.
Diagnostic properties include the type, method, and results. The type of Diagnostic refers to a general category of diagnostic techniques, such as histopathology, serology, imaging, molecular, clinical. Within each type, there are specific methods available, such as biopsy, ELISA assays, X-rays, PCR, or the width of fat in a herd, respectively. In the data model, each Diagnostic has a unit of measurement of its outcome that can either be qualitative or quantitative. For example, an agglutination test can report the minimum dilution that causes observable agglutination. A PCR can be reported as presence of bands compatible with the targeted genome sequence or genetic sequence compatible with targeted organism sequencing followed the genetic sequence amplification.The result of the diagnostic is reported as a value of those units (see Results below).
Diagnostic Products as Specimens
In the data model, Diagnostics can produce “Diagnostic Products” that are then considered a Specimen. Diagnostic products as Specimens can be traced with respect to their storage, the quantities available, and any exportation process (See Storage and Shipment below). Furthermore, Specimens from Diagnostics Products can be used as in other Diagnostics (see Specimens above). For example, cDNA used in a new PCR essay. Basically, the data model accepts a new Specimen generated from a Diagnostic Product with the corresponding type and the origin of the Specimen is a Diagnostic instead of one or multiple Source Records or other Specimens (See Specimens above).
Results
Diagnostics return results in the units of quantitative or qualitative value reported for the Diagnostic. For example, the value of a CT score in real-time PCR. Each Diagnostic can have multiple Results depending on the hazards targeted or it assess the hazard presence multiple times.
Each Result of a Diagnostic has a single Interpretation. The Interpretation is the qualification of the specific Result of a Diagnostic test as positive, negative, or inconclusive, based on case definitions for the diagnostic test reported in the Surveillance Activity metadata.
Storage
In the data model, Specimens from all Sources (including Diagnostic Products) and carcasses of Group and Animal Sources can be stored, the site of storage can be tracked, and changes in storage can be traced. The data recorded also allows to track the amount of Specimen currently available versus the original amount and the reason of the differences over time. Every change in the amount of Specimen and every change in the storage site should be recorded as a different Storage process.
Shipments
In the data model, Specimens from all Sources (including Diagnostic Products) and carcasses of Group and Animal Sources can be shipped to another facility. Partial amounts can also be shipped and the volume exported can be tracked as well as the remaining amounts of the original Specimen left at the storage site. Shipments that are currently “in transit” can also be tracked.
Laboratories
The data model includes Laboratory entities. Laboratories can conduct Diagnostics to test for hazards. Laboratory properties include address, manager, name, Laboratory ID, among others (Data Dictionary). It is also possible to store data regarding laboratory capabilities in terms of diagnostic tests and storage, and their certifications (Bio safety levels, etc.)
Interpretation of Specimens
In the data model, a Specimen can receive an Interpretation with respect to the hazards targeted in the corresponding Surveillance Activity based on a case definition for a positive, negative, or undetermined Specimen provided in the Surveillance Activity metadata. The Interpretation follows the Results from the Diagnostics conducted using the Specimen and their interpretations.
For example, it is possible that a single hazard is assessed in a Specimen using three different Diagnostics and two of them are interpreted as positive and the other one as negative. Based on these Results, the hazard status of the Specimen must be interpreted (positive, negative, inconclusive).
Specimens can receive multiple Interpretations if they are used in several Diagnostics targeting different hazards.
Interpretation of Source Records
In the data model, a Source Record can receive an Interpretation with respect to the hazards targeted in the corresponding Surveillance Activity based on a case definition for a positive, negative, or undetermined Source Record provided in the Surveillance Activity metadata. The Interpretation for a specific hazard follows the Results and Interpretations of the Diagnostics conducted using Specimens of the Source Records and the Interpretation of the Source Records.
For example, it is possible that a single hazard is assessed in multiple Specimens of a Source Record using three different Diagnostics. Two of these Specimens are interpreted as positive and the other one as negative. Based on these data, the hazard status of the Source Record must be interpreted (positive, negative, inconclusive).
Source Record can receive multiple Interpretations if multiple hazards are assessed in them.
Complexities
Tagging Units
The structure of the data model could be enough to record the hierchical data. For example, rangers collecting information during patrols (Collection Tracks) in Protected Areas (Locations) at specific points (Events) where dead animals (Source Records) are found at any given time.
However, the data in another Surveillance Activity could have the following data structure:
- Visit (Field Visit)
- Protected area (Location),
- Zones within protected area (Spatial cluster level 1),
- Grid cells within each zone (Spatial cluster level 2),
- Capture site (Event)
- Mist nets at the capture site (Collection Points),
- Bats captured (Source Records)
- Mist nets at the capture site (Collection Points),
- Capture site (Event)
- Grid cells within each zone (Spatial cluster level 2),
- Zones within protected area (Spatial cluster level 1),
- Protected area (Location),
Another example is surveillance of wild animals in a live market. One potential option to structure these data is:
- Visit (Field Visit)
- City (Spatial cluster level 1)
- Neighborhood (Spatial cluster level 2)
- Market (Location)
- Vendor (Spatial cluster level 3)
- Stalls (Spatial cluster level 4)
- Cage (Event)
- Animals (Source Records)
- Rectal Swab (Specimens)
- Animals (Source Records)
- Cage (Event)
- Stalls (Spatial cluster level 4)
- Vendor (Spatial cluster level 3)
- Market (Location)
- Neighborhood (Spatial cluster level 2)
- City (Spatial cluster level 1)
Looking at the examples above, it is straightforward to understand that the units Location, Event, and Collections might not be enough to accommodate the data of all Surveillance Activities and other units and therefore, it is necessary to tag units with standard labels that allow to group them in further units. *Tags** can group units spatially, temporally, or in another way.
The following example adds the categorization of the data by season, no matter the markets visited or how many years the Surveillance Activity lasts:
- Season (Temporal cluster 1)
- Visit (Field Visit)
- City (Spatial cluster level 1)
- Neighborhood (Spatial cluster level 2)
- Market (Location)
- Vendor (Spatial cluster level 3)
- Stalls (Spatial cluster level 4)
- Cage (Event)
- Animals (Source Records)
- Rectal Swab (Specimens)
- Animals (Source Records)
- Cage (Event)
- Stalls (Spatial cluster level 4)
- Vendor (Spatial cluster level 3)
- Market (Location)
- Neighborhood (Spatial cluster level 2)
- City (Spatial cluster level 1)
The spatial Tags can be nested, non-nested, or a combination of both. Similarly, the temporal Tags can be nested, non-nested, or a combination of both.
Finally, the number of categories within each Cluster can also be different across Surveillance Activities. For example, two Surveillance Activities may include grid cells as one of their Clusters, however, the number of grid cells can be different. For example, one Surveillance Activity can include grid cells A to R, whilst the other Surveillance Activity can include grid cells A to W (more categories). In summary:
- Tags are needed to classify units in further categories
- The Tags across Surveillance Activities can be different
- What these Tags grouped across Surveillance Activities can be different
- The number of categories can be different among Tags, within and between Surveillance Activities
- Nested, Non-nested or both types of Tags can be needed
- Spatial, temporal or both types of Tags can be needed
Surveillance Activities
Initially, it was mentioned that “Field Visits, Locations, Events, Sources, Source Records, and Diagnostics usually belong to a single Surveillance Activity. This is the Surveillance Activity that lead to the Field Visits at different Locations to document Events, collect Sources and Specimens, perform Diagnostics for a specific hazard, and interpret the the results fo Diagnostics and hazard presence in Specimens, and Sources.
Furthermore, it was established that “a Surveillance Activity usually includes Field Visits, Locations, Events, Sources, Source Records, and Diagnostics”.
Here are the exceptions to these two general statements and how the data model can handle them.
Surveillance Activity contains only Specimens and Diagnostics
This can happen when Surveillance Activity 2 uses Specimens collected as part of Surveillance Activity 1 to test them again for the same or different hazard. For example, Specimens from bats collected ten years ago will be used in a new study to test them for SARS-CoV-2. The Specimen belongs to both Surveillance Activities. The Diagnostics are the only new entity generated as part of Surveillance Activity 2, and it is clear that Specimens from Surveillance Activity 1 were used in Surveillance Activity 2.
Surveillance Activity contains only Source Records, Specimens, Diagnostics, and Diagnosis
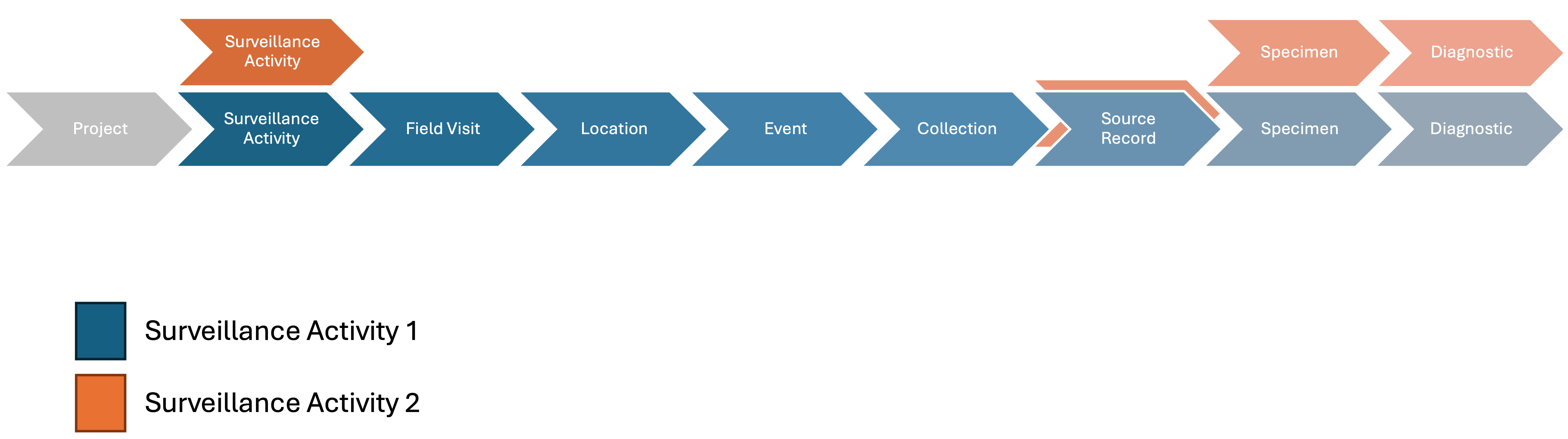
This can happen when Surveillance Activity 2 generates new Specimens from carcasses of Source Records collected as part of Surveillance Activity 1 and tests these new Specimens for a hazard. For example, carcasses of bats collected ten years ago will be used in a new study, Surveillance Activity 2, to get new Specimens and test them for SARS-CoV-2. The Source Records belong to both Surveillance Activities. The new Specimens and Diagnostics are the only new entities generated as part of Surveillance Activity 2, and it is clear that Source Records (the carcasses in this particular case) from Surveillance Activity 1 were used in Surveillance Activity 2.
Same Source in different Surveillance Activities
The health of the same animal could be assessed under different Surveillance Activities. For example, a ranger finds a sick animal in a specific date and assess it health and potential cause of disease as part of Surveillance Activity 1. Then, the animal is taken to a rehabilitation center where its health is assessed again at admission and samples are collected for testing as part of Surveillance Activity 2. Finally, the animal health at the rehabilitation center is assessed again three months later and samples are collected again in order to learn what pathogens are circulating within the facility as part of Surveillance Activity 3.
In this case there are three potential Projects, Surveillance Activities, Field Visits, Events, and Collections Points independently of each other. There are two Locations. The Animal Source belongs to three Surveillance Activities. The Source Records and Specimens belong to a single Surveillance Activity just like the Diagnostics completed in Surveillance in Surveillance Activity 2 and 3.
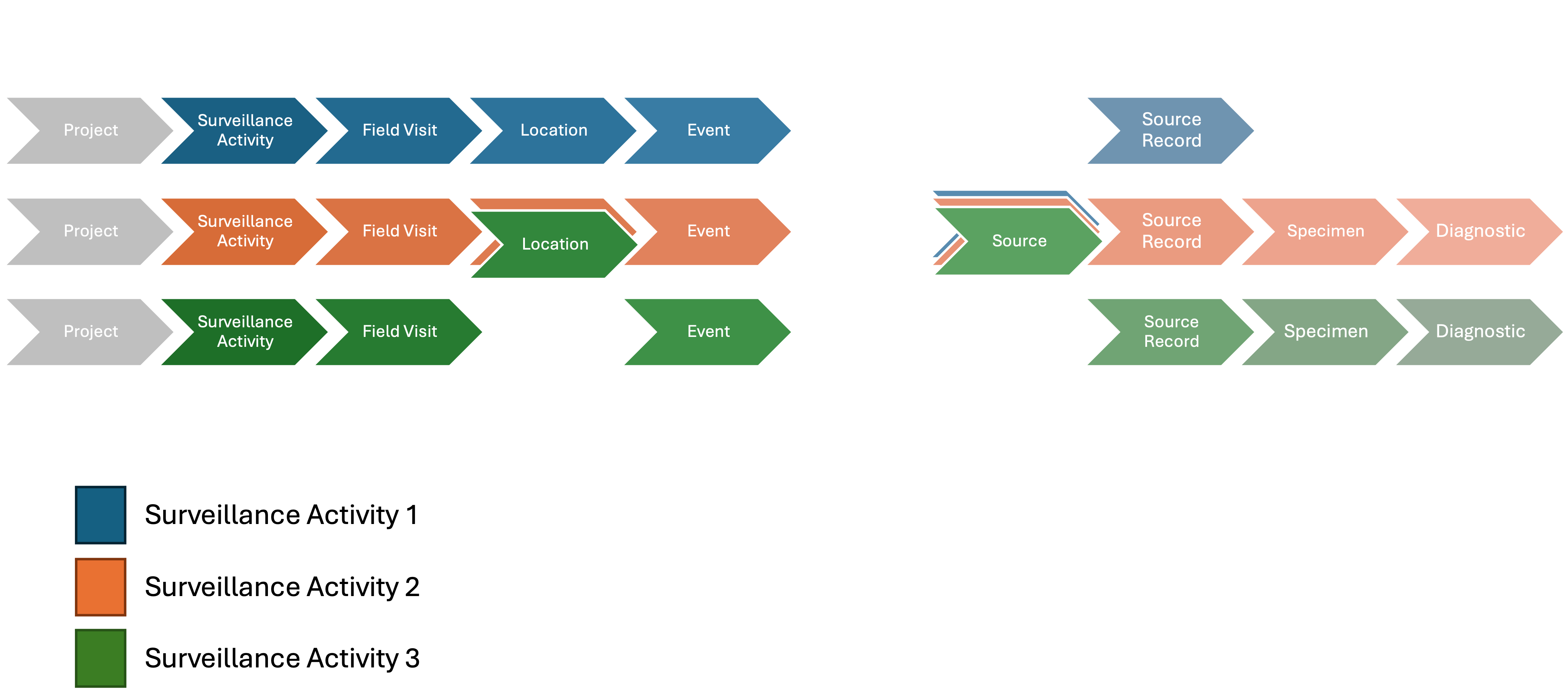
Therefore, Surveillance Activities can be interconnected through common units or the reuse of units generated by previous Surveillance Activities.
Sources in Surveillance Activity but not present in any Event
Studies that track marked individuals over time could succeed in capturing all individually identified animals, so all these Sources are included as Source Records at least once during the study period. However, it is also possible that only a proportion of the individualized Sources are captured or sight during the Surveillance Activity, so these Sources are not present as Source Record of any Event. For the second possibility, it is still important to track the Sources that were part of the sampling frame and censoring them when they were nor part of any Event.
For this reason, the data model can assigned a Surveillance Activity to a Source without including any Field Visits, Location, Event, or Collection Point. When the study is completed, it will be possible to identify all the Sources involved in the study including those that were never captured (Source 3 in the figure below).
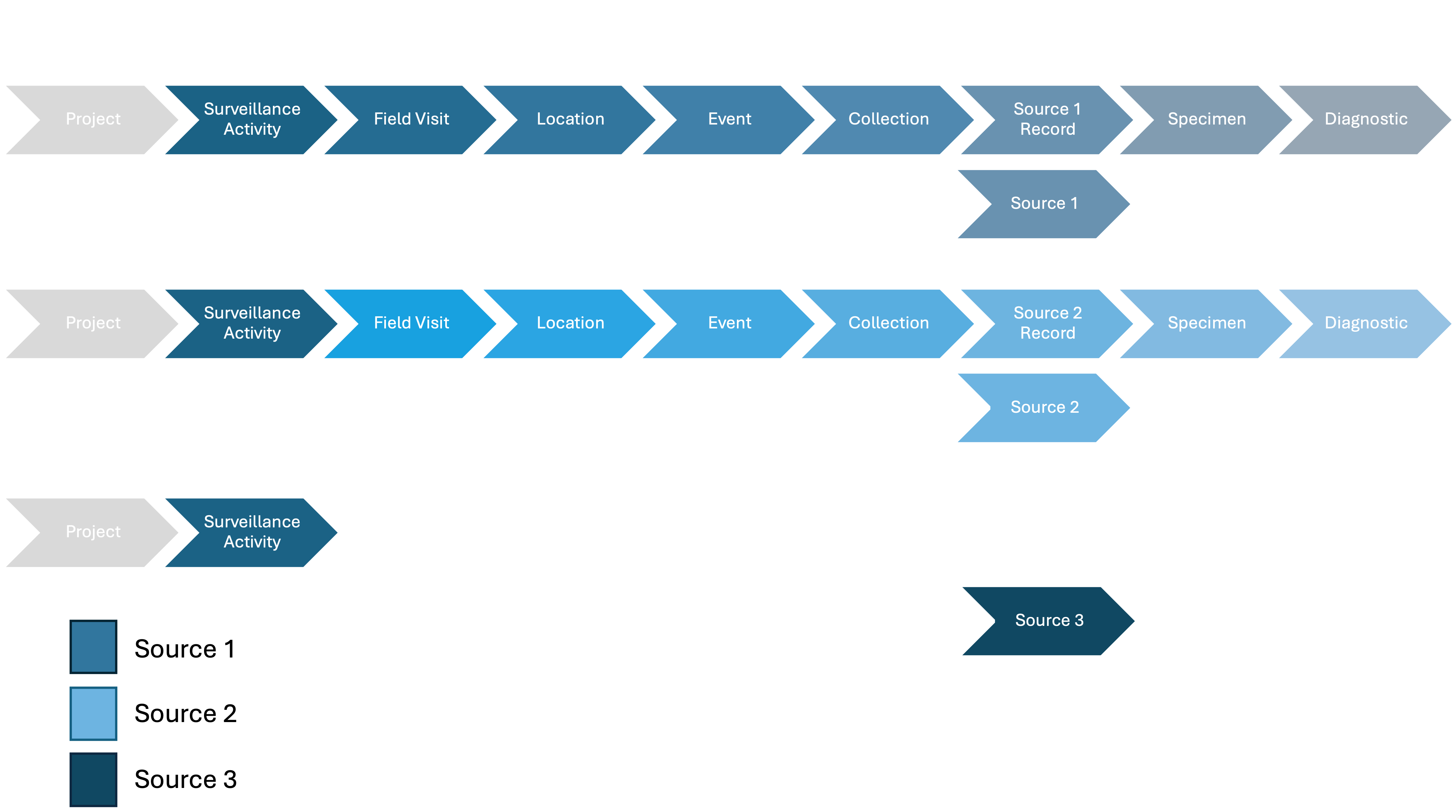
Outbreak Investigation
An outbreak can contains elements associated with two or more Surveillance Activities. For example, the first detection of the outbreak could have been done by a citizen or by rangers patrolling a protected area. The Field Visit, Location, Event, Collection Point (if any), Source Records, and Specimens collected when the outbreak is discovered belong to the “Citizen Science” or “Ranger Patrol” Surveillance Activity. However, they also belong to the outbreak investigation Surveillance Activity, together with Field Visits up to the Diagnostics associated exclusively with the outbreak investigation.
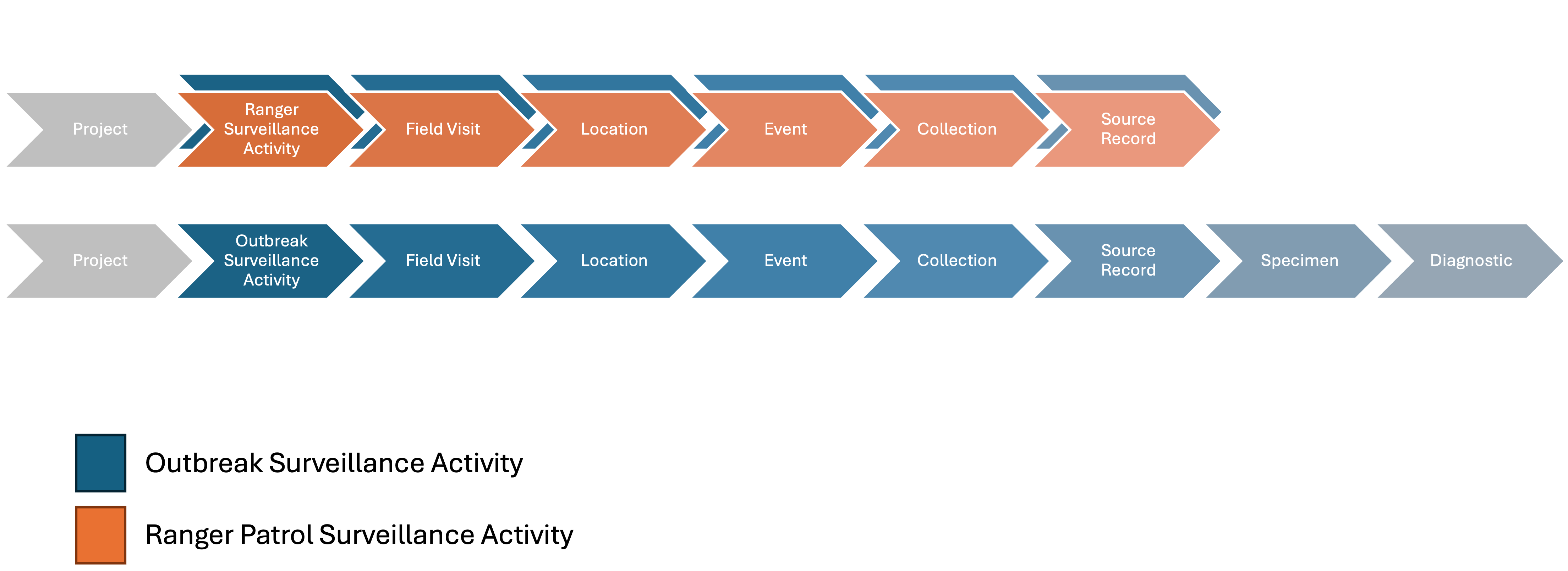
From Group Source to Animal Source
The data model can handle the conversion of an individual in a Group Source into an Animal Source. Animal Sources have a property to identify them as previous membership in a Group Source.
The decision to convert an individual of a Group Source into an Animal Source could occur because: i) it was decided to start recording animals of a Group Source at the individual level including their Specimens and Diagnostics, ii) field Necropsies are conducted with the carcasses of animals that belong to a Group Source (Necropsy data in the data model are linked to individual animals only), iii) all or some carcasses of a Group Source are collected and stored or exported (carcass data in the data model are linked to individual animals only), and iv) Necropsies are conducted with the carcasses of animals belonging to a Group Source in a facility and again, necropsy data in the data model are linked to individual animals only. The data model accepts Diagnostics conducted directly on a Group Source or Diagnostics conducted using Group Source Specimens, such as testing feces taken from a cage occupied by 2 animals of the same species. Therefore, in these cases, it is not necessary to convert individuals of a Group Source to Animal Sources.
In the first case, all or some of the animals of a Group Source could start being recorded at the individual level. If only some of the animals of a Group Source are recorded at the individual level, then it is possible to have an Event at time t with Group Source Records and Animal Source Records of animals that belonged to the Group Source at time t’ that are considered Animal Sources at time t. The Group Source Record does not include the animals that are converted to Animal Sources since time t. Therefore, the count of individuals in the Group Source Record must not include the new Animal Sources. It is also possible that starting at time t in a new Field Visit, all the animals of a Group Source are recorded as Animal Sources. In this case there are no more records of the Group Source after time t-1.
In the second case, a Necropsy conducted with the carcass of an animal of a Group Source leads to the same process explained in the previous paragraph. The animal must be documented at the individual level because, in the data model, Necropsy data are tied to Animal Sources only.
Lastly, the collection of a carcass of an animal of a Group Source and potential subsequent Necropsies also leads to the same process. The animal of a Group Source must be documented at the individual level because, in the data model, *carcass and Necropsy data are tied to Animal Sources only**.
It is important to highlight that: conversion from an individual of a Group Source to Animal Source is complete and irreversible. This means that new Animal Source Record cannot be included in the count of animals in a Group Source, Animal Sources cannot be converted back to be part of the Group Source, and Specimens of the new Animal Sources are not Specimens of the original Group Source. Secondly, when it is decided that animals in a Group Source will be documented as Animal Sources there is a change of methodology and, most likely, the new Animal Sources should receive a new and single Surveillance Activity. It is possible to keep a connection between these Surveillance Activities in the data model (between the Group Source only one and the Groups Source - Animal Sources one). Animal Sources have a property to link them to their original Group Source also. It is possible that the original Surveillance Activity methodology included an eventual change in the recording of animals in a Group Source as Animal Sources and, consequently, only a common unique Surveillance Activity is enough.